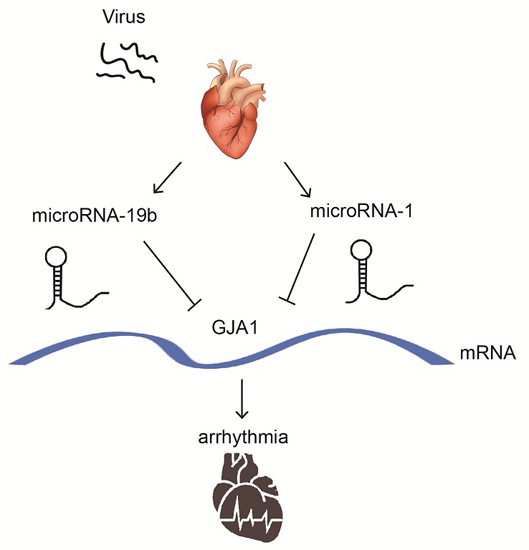
MicroRNA-19b Downregulates Gap Junction Protein Alpha1 and Synergizes with MicroRNA-1 in Viral Myocarditis
Abstract
:1. Introduction
2. Results
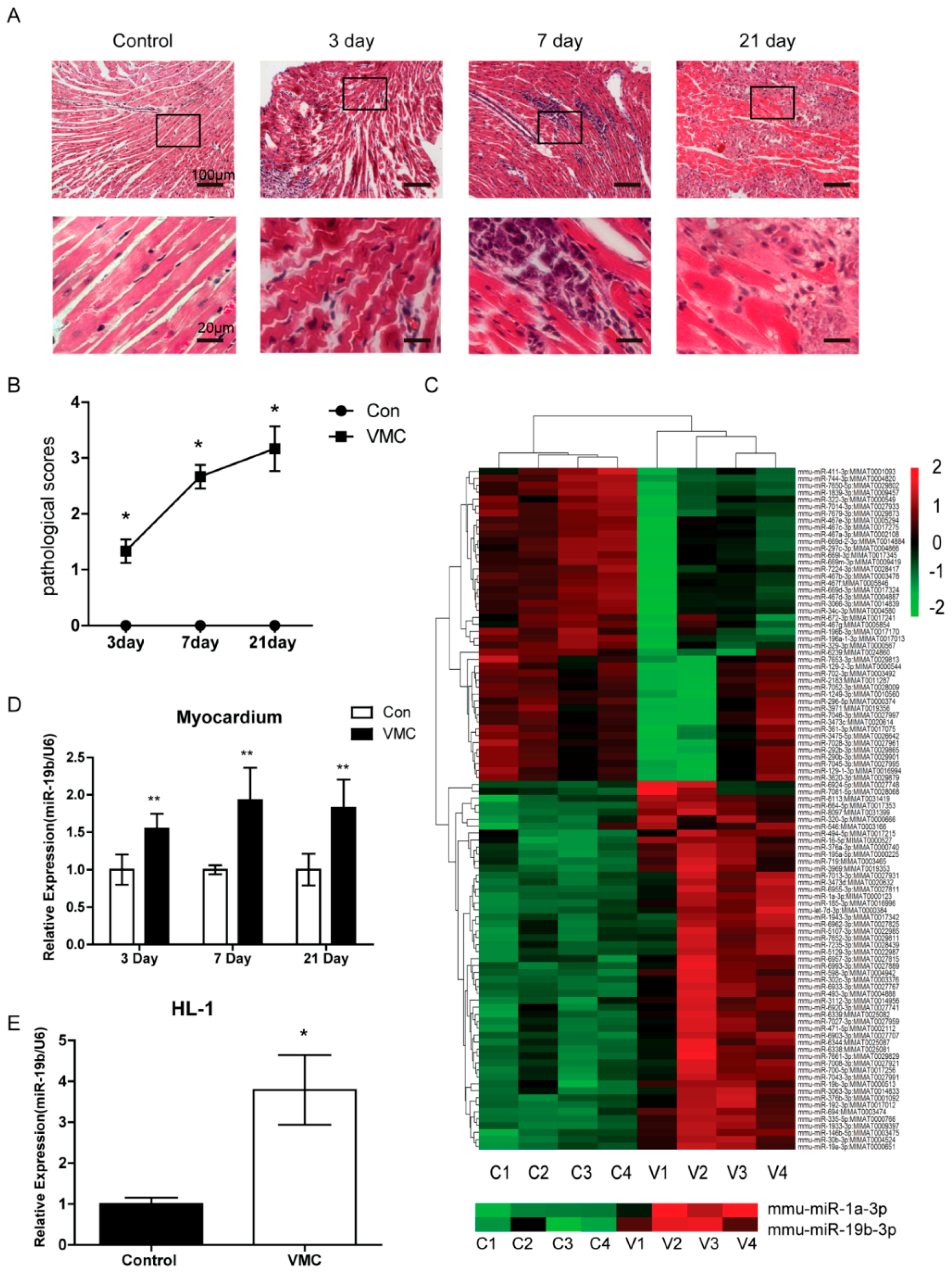
2.1. CVB3 Infection Triggered Significant Myocardial Lesions
2.2. miR-19b Was Upregulated in VMC Model in Vivo and in Vitro
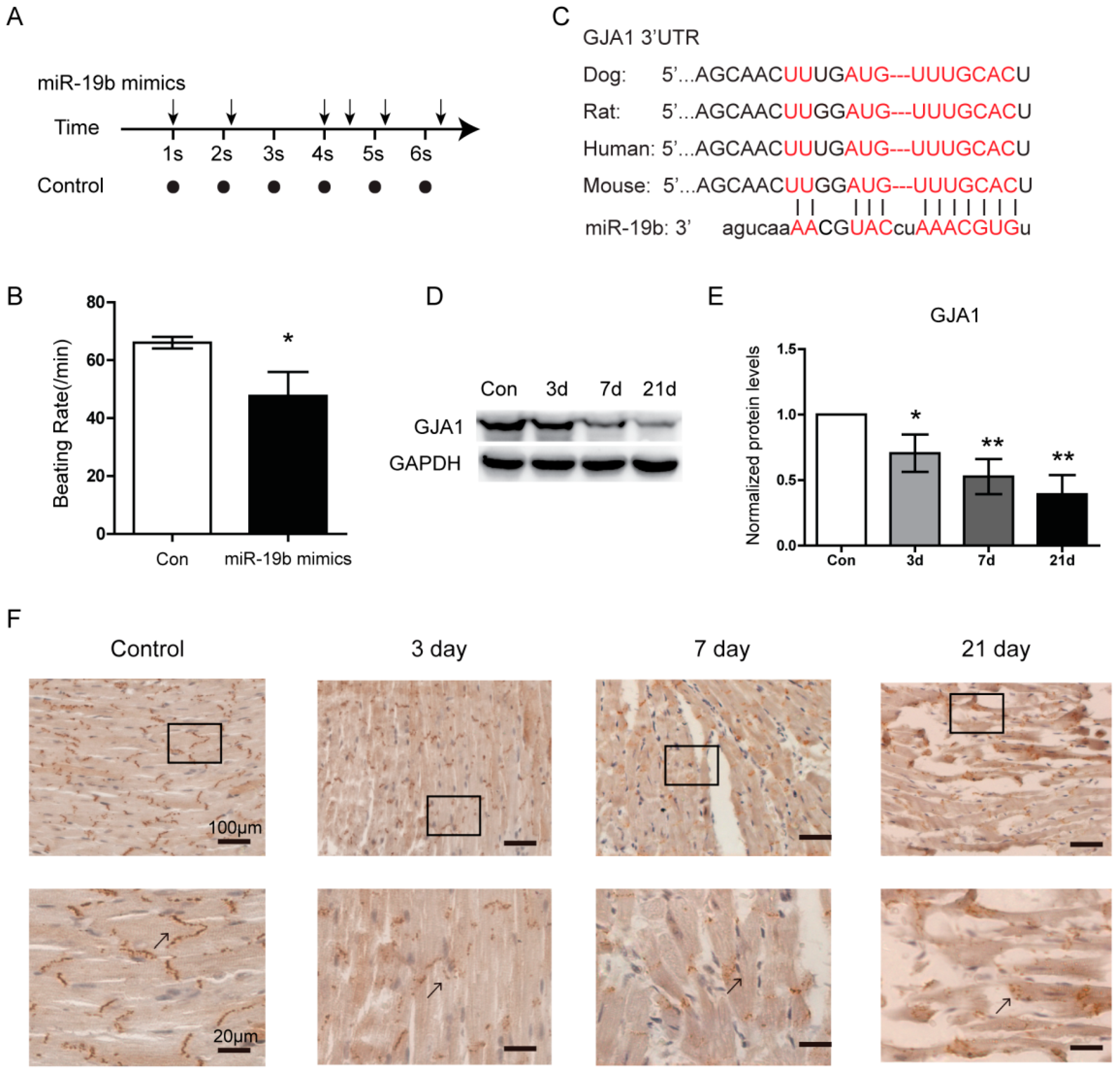
2.3. Overexpression of miR-19b Resulted in Irregular Beating Patterns in hiPSCs-CMs
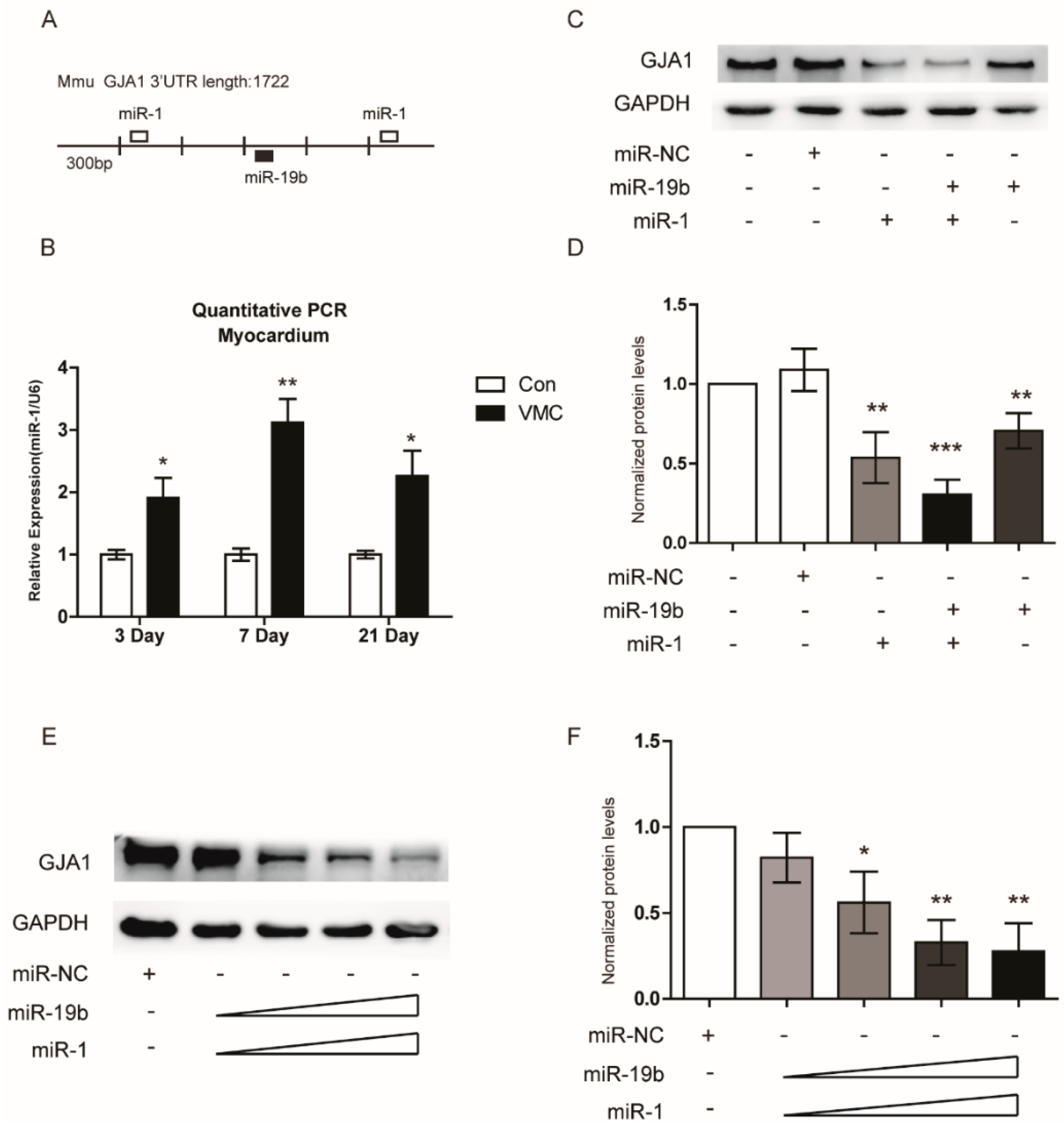
2.4. GJA1 Was Predicted to Be Targeted by miR-19b and Was Downregulated in VMC
2.5. miR-19b Directly Regulated the Expression of GJA1 in Vitro
2.6. Re-Expression of GJA1 Rescued the miR-19b-Mediated Irregular Beating in hiPSCs-CMs
2.7. miR-19b Cooperated with miR-1 to Regulate the GJA1 Expression
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Histopathological Analysis and Myocardial Pathological Scoring
4.3. Immunohistochemistry (IHC)
4.4. Western Blot Analysis
4.5. RNA Analysis
4.6. miRNA Microarray Analysis
4.7. Luciferase Activity Assay
4.8. Cell Culture and Transfection in Vitro
4.9. Statistics
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| VMC | Viral myocarditis |
| MiRNAs | MircroRNAs |
| GJA1 | Gap junction protein alpha 1 |
| hiPSCs-CMs | human cardiomyocytes derived from the induced pluripotent stem cells |
| CVB3 | Coxsackievirus B3 |
| 3’-UTR | 3’-untranslated region |
| TCID50 | 50% tissue culture infectious dose |
| i.p. | Intraperitoneally |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
References
- Cooper, L.J.; Keren, A.; Sliwa, K.; Matsumori, A.; Mensah, G.A. The global burden of myocarditis: A systematic literature review for the Global Burden of Diseases, Injuries, and Risk Factors 2010 study. Glob. Heart 2014, 9, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.; Liu, P.P.; Cooper, L.J. Myocarditis. Lancet 2012, 379, 738–747. [Google Scholar] [CrossRef]
- Blauwet, L.A.; Cooper, L.T. Myocarditis. Prog. Cardiovasc. Dis. 2010, 52, 274–288. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L.J. Myocarditis. N. Engl. J. Med. 2009, 360, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Esfandiarei, M.; McManus, B.M. Molecular biology and pathogenesis of viral myocarditis. Annu. Rev. Pathol. 2008, 3, 127–155. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, G.G.; van Eyk, J.E.; Tomaselli, G.F. Mechanisms of gap junction traffic in health and disease. J. Cardiovasc. Pharmacol. 2009, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Michela, P.; Velia, V.; Aldo, P.; Ada, P. Role of connexin 43 in cardiovascular diseases. Eur. J. Pharmacol. 2015, 768, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Van Rijen, H.V.; Eckardt, D.; Degen, J.; Theis, M.; Ott, T.; Willecke, K.; Jongsma, H.J.; Opthof, T.; de Bakker, J.M. Slow conduction and enhanced anisotropy increase the propensity for ventricular tachyarrhythmias in adult mice with induced deletion of connexin43. Circulation 2004, 109, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Akar, F.G.; Spragg, D.D.; Tunin, R.S.; Kass, D.A.; Tomaselli, G.F. Mechanisms underlying conduction slowing and arrhythmogenesis in nonischemic dilated cardiomyopathy. Circ. Res. 2004, 95, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Lin, H.; Xiao, J.; Lu, Y.; Luo, X.; Li, B.; Zhang, Y.; Xu, C.; Bai, Y.; Wang, H.; Chen, G.; Wang, Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007, 13, 486–491. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Corsten, M.; Heggermont, W.; Papageorgiou, A.P.; Deckx, S.; Tijsma, A.; Verhesen, W.; van Leeuwen, R.; Carai, P.; Thibaut, H.J.; Custers, K.; et al. The microRNA-221/-222 cluster balances the antiviral and inflammatory response in viral myocarditis. Eur. Heart J. 2015. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.F.; Ding, Y.J.; Shen, Y.W.; Xue, A.M.; Xu, H.M.; Luo, C.L.; Li, B.X.; Liu, Y.L.; Zhao, Z.Q. MicroRNA-1 represses Cx43 expression in viral myocarditis. Mol. Cell. Biochem. 2012, 362, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.T. miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008, 133, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, M.; Mummery, C.L.; Wilde, A.A.; Bezzina, C.R.; Verkerk, A.O. Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias. Front. Physiol. 2012, 3, 346. [Google Scholar] [CrossRef] [PubMed]
- Priori, S.G.; Napolitano, C.; Di Pasquale, E.; Condorelli, G. Induced pluripotent stem cell-derived cardiomyocytes in studies of inherited arrhythmias. J. Clin. Investig. 2013, 123, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Baksi, A.J.; Kanaganayagam, G.S.; Prasad, S.K. Arrhythmias in viral myocarditis and pericarditis. Card Electrophysiol. Clin. 2015, 7, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zhang, H.; Wu, S. Immunomodulation by atorvastatin upregulates expression of gap junction proteins in coxsackievirus B3 (CVB3)-induced myocarditis. Inflamm. Res. 2010, 59, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Gutstein, D.E.; Morley, G.E.; Tamaddon, H.; Vaidya, D.; Schneider, M.D.; Chen, J.; Chien, K.R.; Stuhlmann, H.; Fishman, G.I. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circ. Res. 2001, 88, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Del, R.S.; Moscato, S.; Bianchi, F.; Morales, M.A.; Dolfi, A.; Burchielli, S.; Cabiati, M.; Mattii, L. Altered expresAltered expression of connexin 43 and related molecular partners in a pig model of left ventricular dysfunction with and without dipyrydamole therapy. Pharmacol. Res. 2015, vol. 92–101. [Google Scholar]
- Severs, N.J.; Coppen, S.R.; Dupont, E.; Yeh, H.I.; Ko, Y.S.; Matsushita, T. Gap junction alterations in human cardiac disease. Cardiovasc. Res. 2004, 62, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Mitcheson, J.S.; Hancox, J.C.; Levi, A.J. Cultured adult cardiac myocytes: Future applications, culture methods, morphological and electrophysiological properties. Cardiovasc. Res. 1998, 39, 280–300. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Claycomb, W.C.; Lanson, N.J.; Stallworth, B.S.; Egeland, D.B.; Delcarpio, J.B.; Bahinski, A.; Izzo, N.J. HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA 1998, 95, 2979–2984. [Google Scholar] [CrossRef] [PubMed]
- Sinnecker, D.; Laugwitz, K.L.; Moretti, A. Extending human induced pluripotent stem cell technology to infectious diseases: New model for viral myocarditis. Circ. Res. 2014, 115, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Corsten, M.F.; Papageorgiou, A.; Verhesen, W.; Carai, P.; Lindow, M.; Obad, S.; Summer, G.; Coort, S.L.; Hazebroek, M.; van Leeuwen, R.; et al. MicroRNA profiling identifies microRNA-155 as an adverse mediator of cardiac injury and dysfunction during acute viral myocarditis. Circ. Res. 2012, 111, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.F.; Ding, Y.J.; Zhang, Z.X.; Wang, Z.F.; Luo, C.L.; Li, B.X.; Shen, Y.W.; Tao, L.Y.; Zhao, Z.Q. MicroRNA‑21 regulation of the progression of viral myocarditis to dilated cardiomyopathy. Mol. Med. Rep. 2014, 10, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Song, D.W.; Ryu, J.Y.; Kim, J.O.; Kwon, E.J.; Kim, D.H. The miR-19a/b family positively regulates cardiomyocyte hypertrophy by targeting atrogin-1 and MuRF-1. Biochem. J. 2014, 457, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hu, X.; Zhu, J.; Zhu, C.; Zhu, S.; Liu, X.; Xu, J.; Han, S.; Yu, Z. Overexpression of miR-19b impairs cardiac development in zebrafish by targeting CTNNB1. Cell. Physiol. Biochem. 2014, 33, 1988–2002. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, Y.C.; Chen, Y.; Xiang, Y.; Shen, C.X.; Li, Y.G. The role of miR-19b in the inhibition of endothelial cell apoptosis and its relationship with coronary artery disease. Sci. Rep. 2015, 5, 15132. [Google Scholar] [CrossRef] [PubMed]
- Danielson, L.S.; Park, D.S.; Rotllan, N.; Chamorro-Jorganes, A.; Guijarro, M.V.; Fernandez-Hernando, C.; Fishman, G.I.; Phoon, C.K.; Hernando, E. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013, 27, 1460–1467. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Sun, L.Y.; Zhang, S.C.; Wei, R.; Xie, F.; Liu, J.; Yan, Y.; Duan, M.J.; Sun, L.L.; Sun, Y.H.; et al. MicroRNA-23a participates in estrogen deficiency induced gap junction remodeling of rats by targeting GJA1. Int. J. Biol. Sci. 2015, 11, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Osbourne, A.; Calway, T.; Broman, M.; McSharry, S.; Earley, J.; Kim, G.H. Downregulation of connexin43 by microRNA-130a in cardiomyocytes results in cardiac arrhythmias. J. Mol. Cell. Cardiol. 2014, 74, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Ares-Carrasco, S.; Picatoste, B.; Camafeita, E.; Carrasco-Navarro, S.; Zubiri, I.; Ortiz, A.; Egido, J.; Lopez, J.A.; Tunon, J.; Lorenzo, O. Proteome changes in the myocardium of experimental chronic diabetes and hypertension: Role of PPARα in the associated hypertrophy. J. Proteom. 2012, 75, 1816–1829. [Google Scholar] [CrossRef] [PubMed]
- Knollmann, B.C. Induced pluripotent stem cell-derived cardiomyocytes: Boutique science or valuable arrhythmia model? Circ. Res. 2013, 112, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Fukuoka, M.; Li, G.; Liu, Y.; Chen, M.; Konviser, M.; Chen, X.; Opavsky, M.A.; Liu, P.P. Regulatory T cells protect mice against coxsackievirus-induced myocarditis through the transforming growth factor β-coxsackie-adenovirus receptor pathway. Circulation 2010, 121, 2624–2634. [Google Scholar] [CrossRef] [PubMed]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.M.; et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 2014, 11, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Ke, Q.; Li, L.; Cai, B.; Liu, C.; Yang, Y.; Gao, Y.; Huang, W.; Yuan, X.; Wang, T.; Zhang, Q.; et al. Connexin 43 is involved in the generation of human-induced pluripotent stem cells. Hum. Mol. Genet. 2013, 22, 2221–2233. [Google Scholar] [CrossRef] [PubMed]


| Score | Pathological Display |
|---|---|
| 0 | no inflammatory infiltrates |
| 1 | small foci of inflammatory cells between myocytes and the total inflammatory infiltrate area was less than 5% of the cross section |
| 2 | the total inflammatory infiltrate area was between 5% and 10% |
| 3 | inflammatory infiltrate area was between 10% and 25% |
| 4 | more than 25% inflammatory infiltrates or diffuse fibrosis lesions with necrosis |
| miRNAs | Primer Sequences 5’-3’ |
|---|---|
| mmu-miR-19b-3p | TGTGCAAATCCATGCAAAACTGA |
| mmu-miR-1a-3p | TGGAATGTAAAGAAGTATGTAT |
| U6 Forward | CAAGGATGACACGCAAATTCG |
| U6 Reverse | ACACGCAAATTCGTGAAGC |
| Genes | Primer Sequences 5’-3’ | |
|---|---|---|
| GJA1 | Forward | CACTGAGCCCATCCAAAGA |
| Reverse | TGTACCCAGGAGGAGACATAG | |
| GAPDH | Forward | TGCGACTTCAACAGCAACTC |
| Reverse | ATGTAGGCCATGAGGTCCAC | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, J.; Xue, A.; Li, L.; Li, B.; Li, Y.; Shen, Y.; Sun, N.; Chen, R.; Xu, H.; Zhao, Z. MicroRNA-19b Downregulates Gap Junction Protein Alpha1 and Synergizes with MicroRNA-1 in Viral Myocarditis. Int. J. Mol. Sci. 2016, 17, 741. https://doi.org/10.3390/ijms17050741
Lin J, Xue A, Li L, Li B, Li Y, Shen Y, Sun N, Chen R, Xu H, Zhao Z. MicroRNA-19b Downregulates Gap Junction Protein Alpha1 and Synergizes with MicroRNA-1 in Viral Myocarditis. International Journal of Molecular Sciences. 2016; 17(5):741. https://doi.org/10.3390/ijms17050741
Chicago/Turabian StyleLin, Junyi, Aimin Xue, Liliang Li, Beixu Li, Yuhua Li, Yiwen Shen, Ning Sun, Ruizhen Chen, Hongfei Xu, and Ziqin Zhao. 2016. "MicroRNA-19b Downregulates Gap Junction Protein Alpha1 and Synergizes with MicroRNA-1 in Viral Myocarditis" International Journal of Molecular Sciences 17, no. 5: 741. https://doi.org/10.3390/ijms17050741