Is the Efficiency of RNA Silencing Evolutionarily Regulated?
Abstract
:1. Introduction
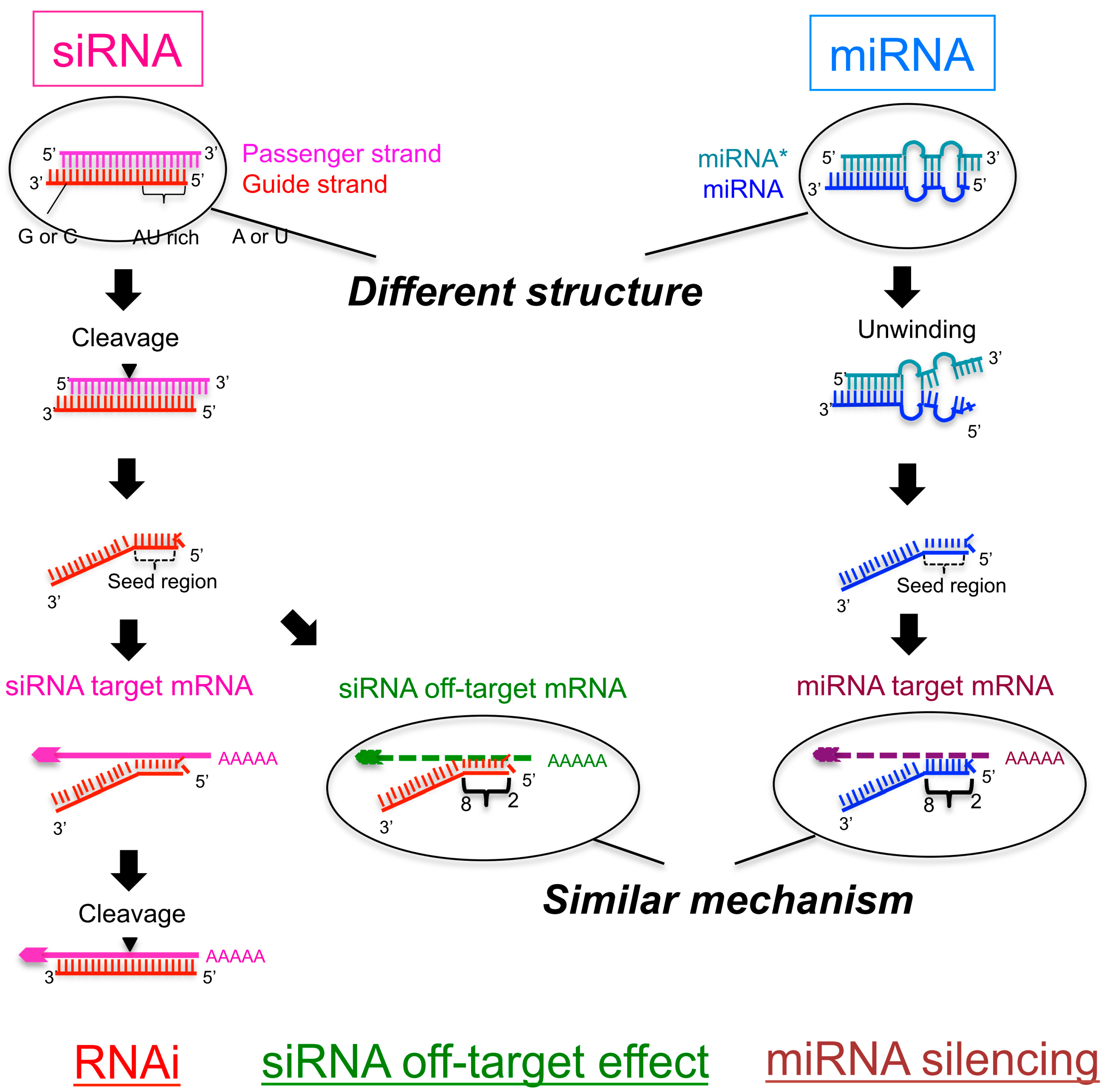
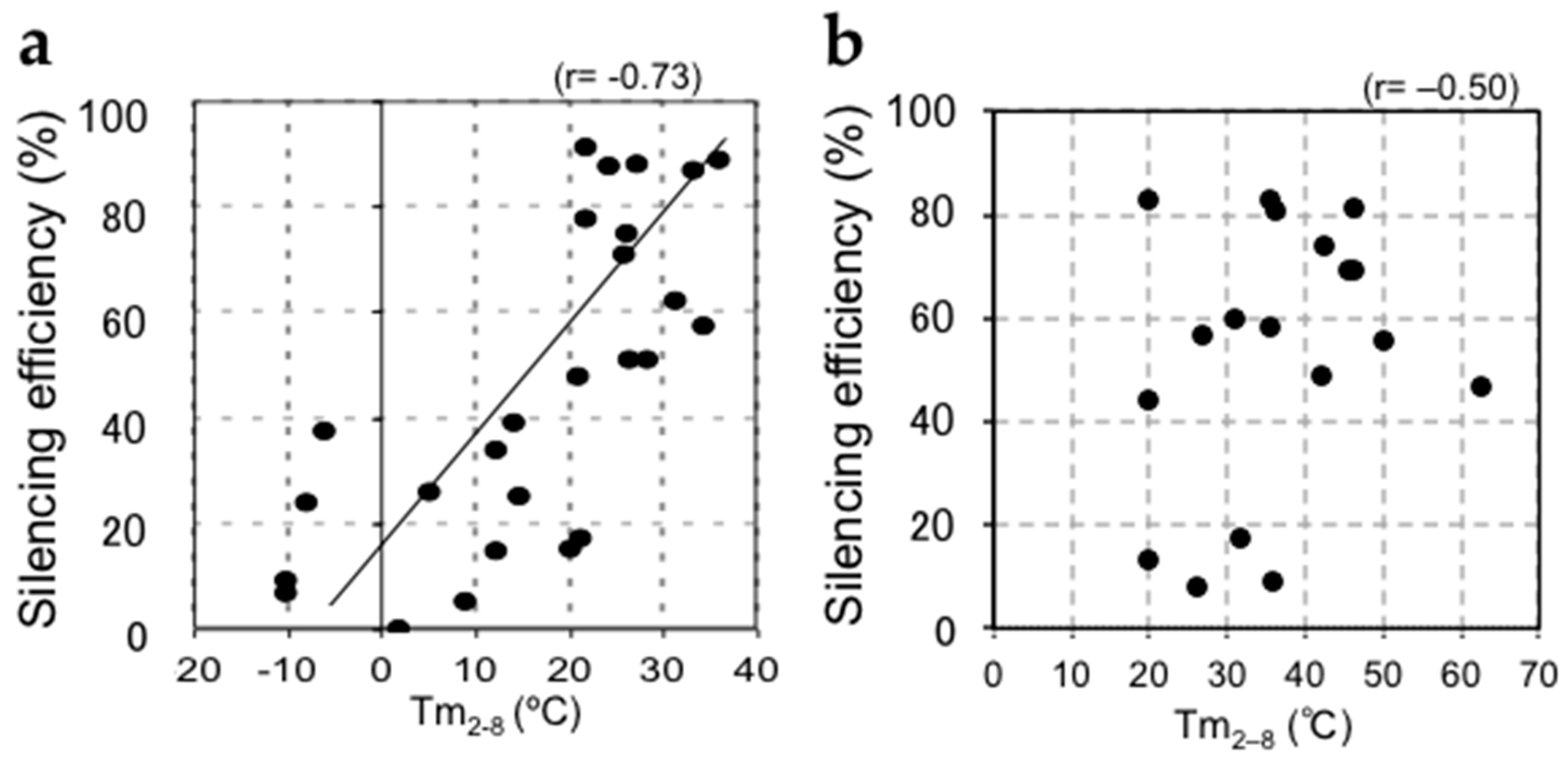
2. The Silencing Efficiency of siRNAs on Off-Target mRNAs Is Regulated by the Stability of Base Pairing between the siRNA Seed Region and Off-Target mRNA
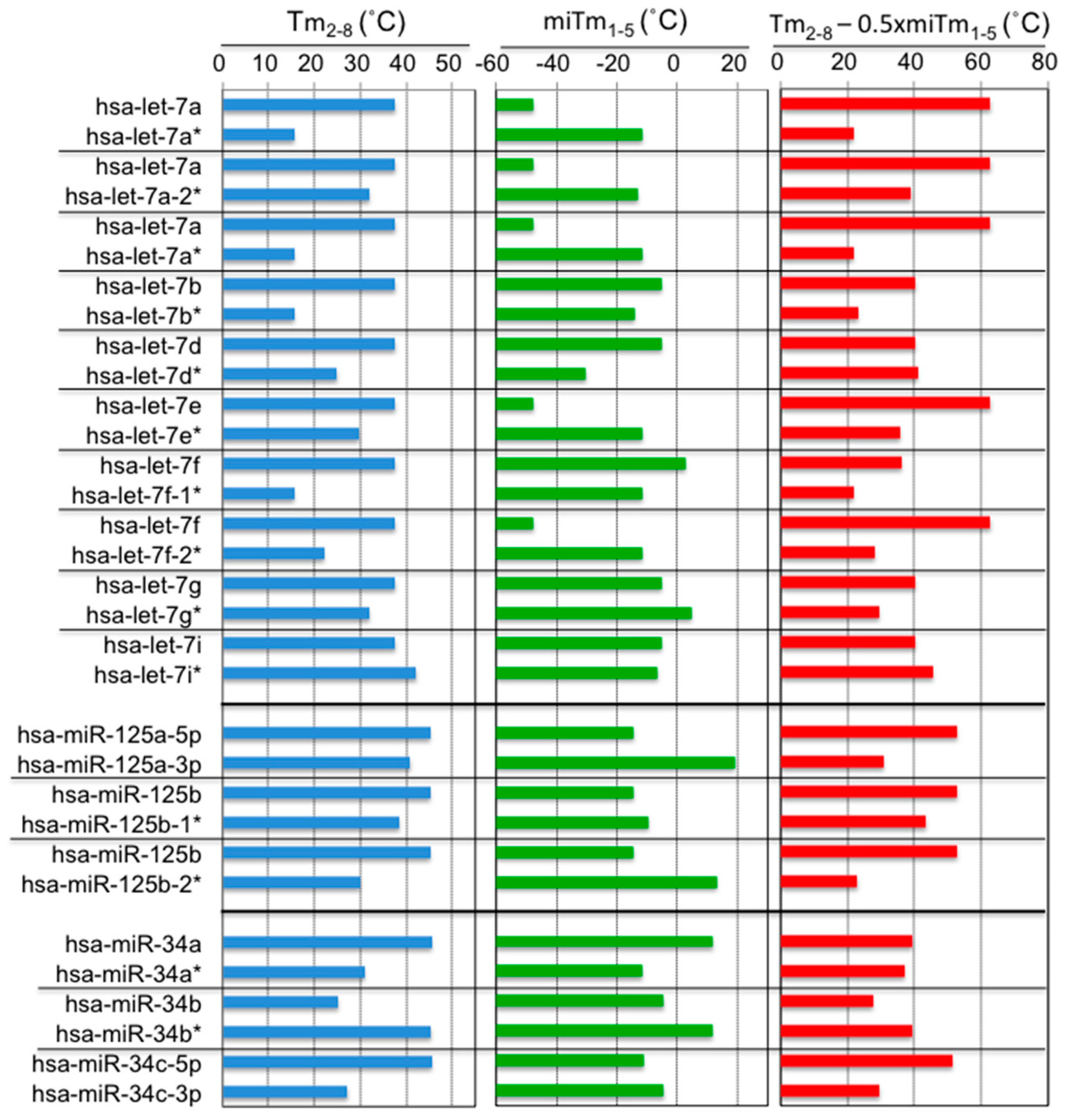
3. The Efficiency of Silencing of Target mRNAs by miRNAs Is Regulated by the Stability of Base Pairing in the 5′ Region of the miRNA and that between the miRNA Seed Region and Target mRNAs
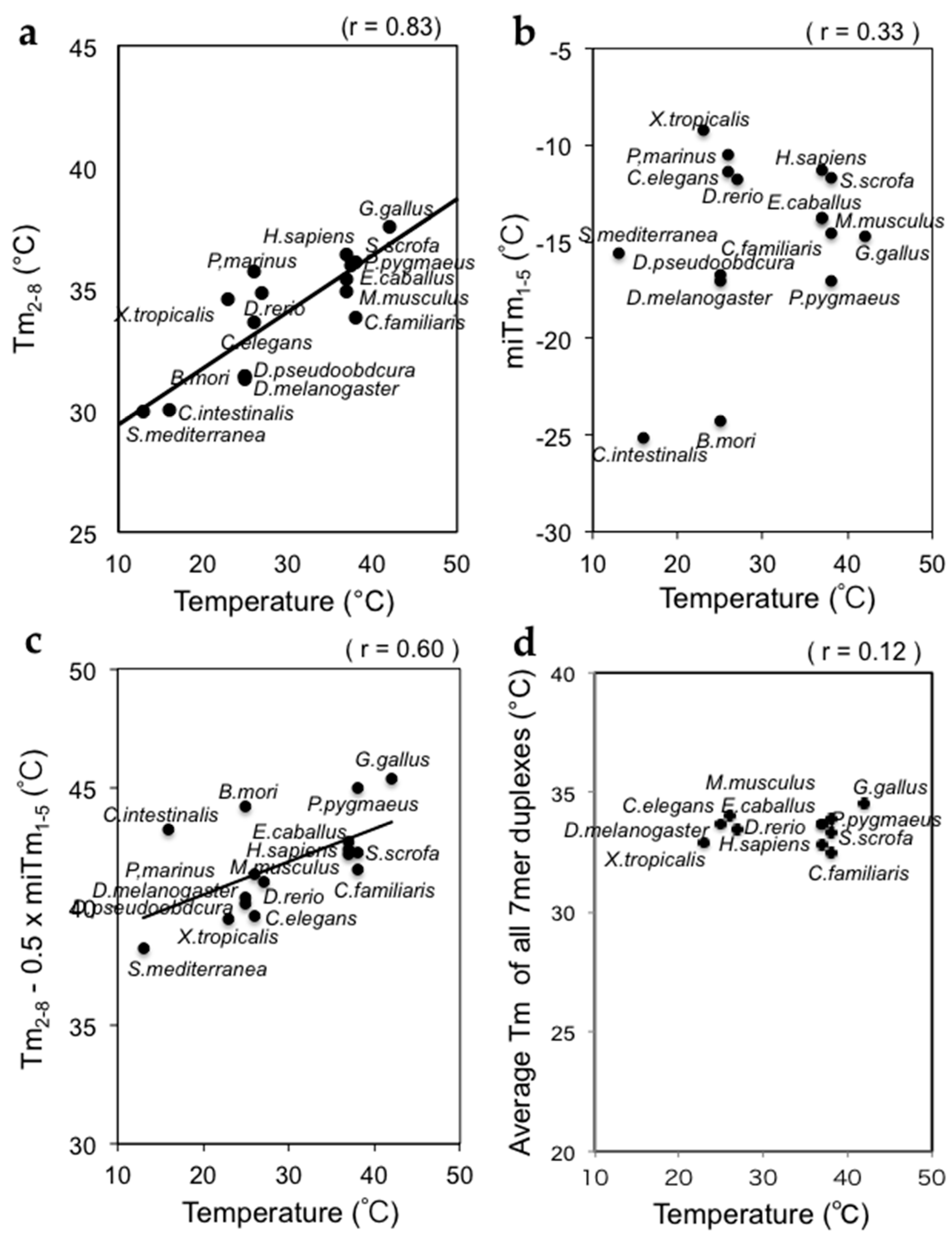
4. The Base Pairing Stabilities of miRNAs in Various Species are Strongly Correlated with Body Temperature
5. Perspectives
Acknowledgments
Conflicts of Interest
Abbreviations
| siRNA | small interfering RNA |
| miRNA | microRNA |
| UTR | untranslated region |
| ncRNA | non-coding RNA |
| pri-miRNA | primary miRNA |
| pre-miRNA | precursor miRNA |
| AGO | Argonaute |
| RISC | RNA-induced silencing complex |
| CDS | coding sequence |
| RNAi | RNA interference |
| Tm | melting temperature |
References
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Witkos, T.M.; Koscianska, E.; Krzyzosiak, W.J. Practical aspects of microRNA target prediction. Curr. Mol. Med. 2011, 11, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2001, 21, 4663–4670. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. Sequence requirements for micro RNA processing and function in human cells. RNA 2003, 9, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Matranga, C.; Tomori, Y.; Shin, C.; Bartel, D.P.; Zamore, P.S. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Tsukumo, H.; Nagami, T.; Siomi, H.; Siomi, M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Gen. Dev. 2005, 19, 2837–2848. [Google Scholar] [CrossRef] [PubMed]
- Leuschner, P.J.; Ameres, S.L.; Kueng, S.; Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006, 7, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2009, 17, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Ruan, X.; Anderson, M.G.; McDowell, J.A.; Kroeger, P.E.; Resik, S.W.; Shen, Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005, 33, 4527–4535. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sonheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [PubMed]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Gen. Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.; Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001, 20, 6877–6888. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Luhrmann, R.; Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef]
- Nykanen, A.; Haley, B.; Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 2001, 107, 309–321. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvagner, G.; Haley, B.; Zamore, P.D. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 2002, 10, 537–548. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.R.; Pei, Y.; Ma, J.B.; Kuryavyi, V.; Zhadina, M.; Meister, G.; Chen, H.Y.; Dauter, Z.; Tuschl, T.; Patel, D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 2005, 19, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.B.; Yuan, Y.R.; Meister, G.; Pei, Y.; Tuschl, T.; Patel, D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005, 434, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Ui-Tei, K.; Naito, Y.; Nishi, K.; Juni, A.; Saigo, K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008, 36, 7100–7109. [Google Scholar] [CrossRef] [PubMed]
- Ui-Tei, K.; Naito, Y.; Takahashi, F.; Haraguchi, T.; Ohki-Hamazaki, H.; Juni, A.; Ueda, R.; Saigo, K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004, 32, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Kamola, P.J.; Nakano, Y.; Takahashi, T.; Wilson, P.; Ui-Tei, K. The siRNA non-seed region and its target sequences are auxiliary determinants of off-target effects. PLoS Comput. Biol. 2015, 11, 41004656. [Google Scholar] [CrossRef] [PubMed]
- Hibio, N.; Hino, K.; Shimizu, E.; Nagata, Y.; Ui-Tei, K. Stability of miRNA 5′ terminal and seed regions is correlated with experimentally observed miRNA-mediated silencing efficacy. Sci. Rep. 2012, 2, 996. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; SantaLucia, J., Jr.; Burkard, M.E.; Kierzek, R.; Schroeder, S.J.; Jiao, X.; Cox, C.; Turner, D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 1998, 37, 14719–14735. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [PubMed]
- Caygill, E.E.; Johnston, L.A. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 2008, 18, 943–950. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.Y.; Chung, K.H.; Deo, M.; Thompson, R.C.; Turner, D.L. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp. Cell Res. 2008, 31, 2618–2633. [Google Scholar] [CrossRef] [PubMed]
- Takane, K.; Fujishima, K.; Watanabe, Y.; Sato, A.; Saito, N.; Tomita, M.; Kanai, A. Computational prediction and experimental validation of evolutionarily conserved microRNA target genes in bilaterian animals. BMC Genom. 2010, 11, 101. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Lobry, J.R. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 1997, 44, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D.; Merchant, A.R. High guanine-cytosine content is not an adaptation to high temperature: A comparative analysis amongst prokaryotes. Proc. R. Soc. B Biol. Sci. 2001, 268, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Forsdyke, D.R.; Bell, S.J. Purine loading, stem-loops and Chargaff’s second parity rule: A discussion of the application of elementary principles to early chemical observations. Appl. Bioinform. 2004, 3, 3–8. [Google Scholar] [CrossRef]
- Forterre, P. A hot story from comparative genomics: Reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002, 18, 236–237. [Google Scholar] [CrossRef]
- Nakashima, H.; Fukuchi, S.; Nishikawa, K. Compositional changes in RNA, DNA and proteins for bacterial adaptation to higher and lower temperatures. J. Biochem. 2003, 133, 507–513. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ui-Tei, K. Is the Efficiency of RNA Silencing Evolutionarily Regulated? Int. J. Mol. Sci. 2016, 17, 719. https://doi.org/10.3390/ijms17050719
Ui-Tei K. Is the Efficiency of RNA Silencing Evolutionarily Regulated? International Journal of Molecular Sciences. 2016; 17(5):719. https://doi.org/10.3390/ijms17050719
Chicago/Turabian StyleUi-Tei, Kumiko. 2016. "Is the Efficiency of RNA Silencing Evolutionarily Regulated?" International Journal of Molecular Sciences 17, no. 5: 719. https://doi.org/10.3390/ijms17050719





