Hairpin RNA Targeting Multiple Viral Genes Confers Strong Resistance to Rice Black-Streaked Dwarf Virus
Abstract
:1. Introduction
2. Results
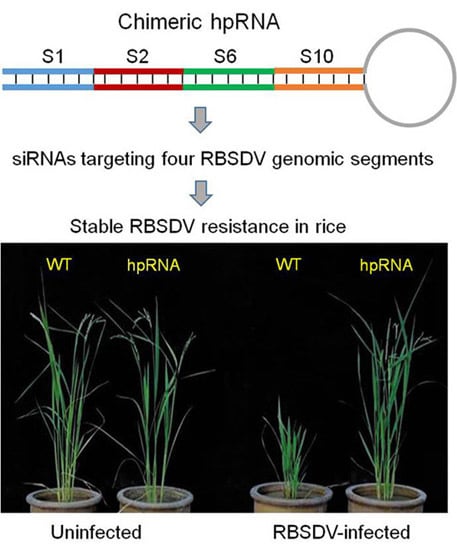
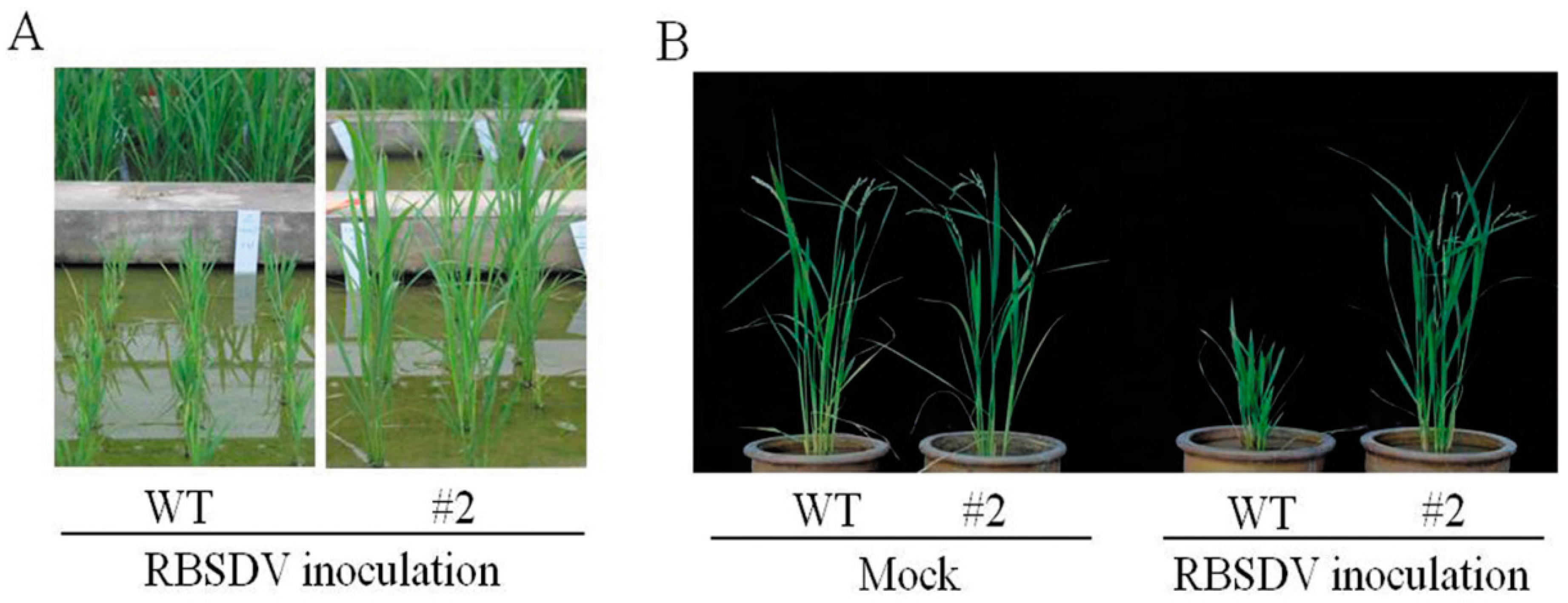
2.1. Rice Plants Transformed with an hpRNA Construct Targeting Four RBSDV Genes Show Strong Resistance to RBSDV
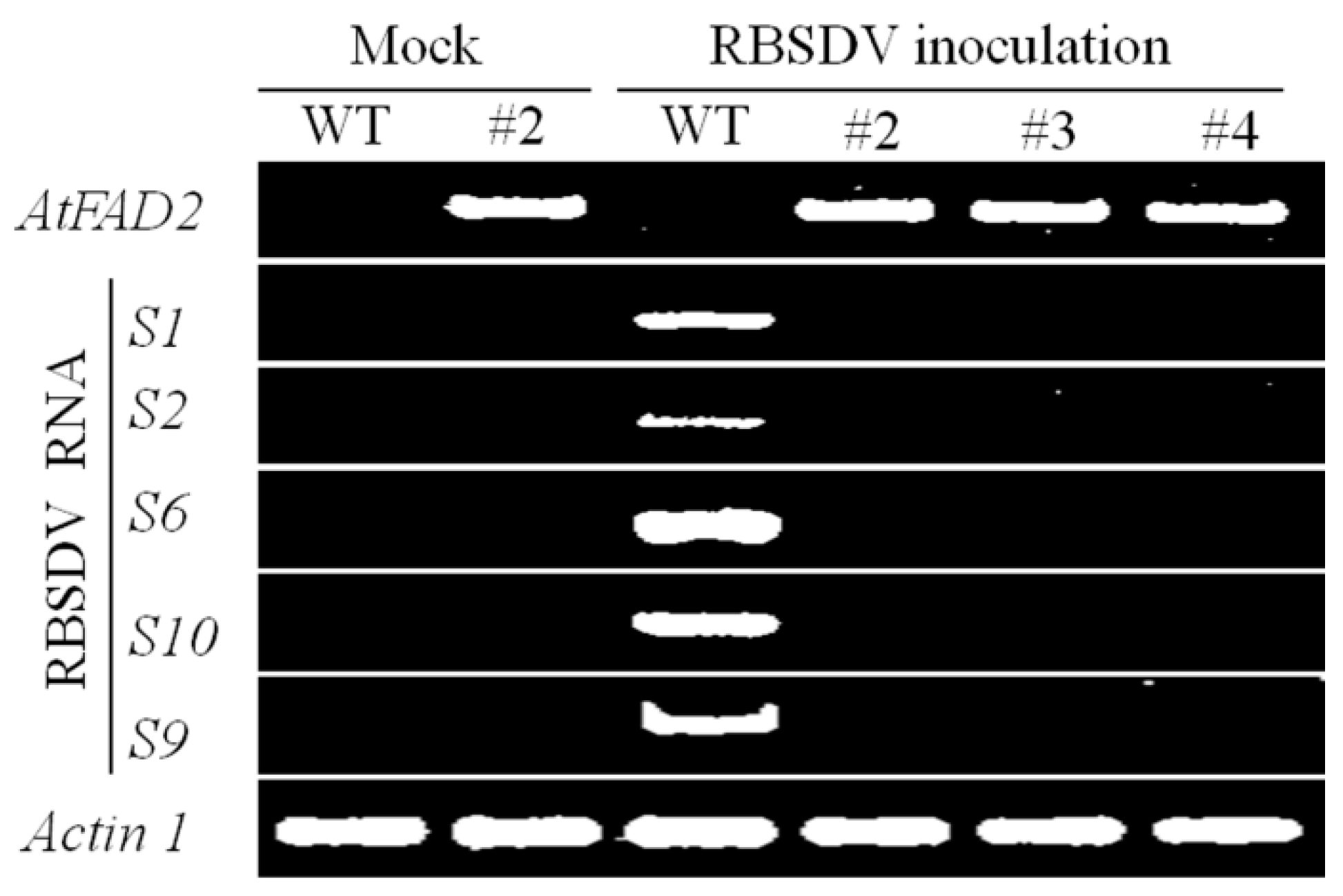
2.2. RBSDV Accumulation Is Inhibited in the hpRNA Transgenic Plants
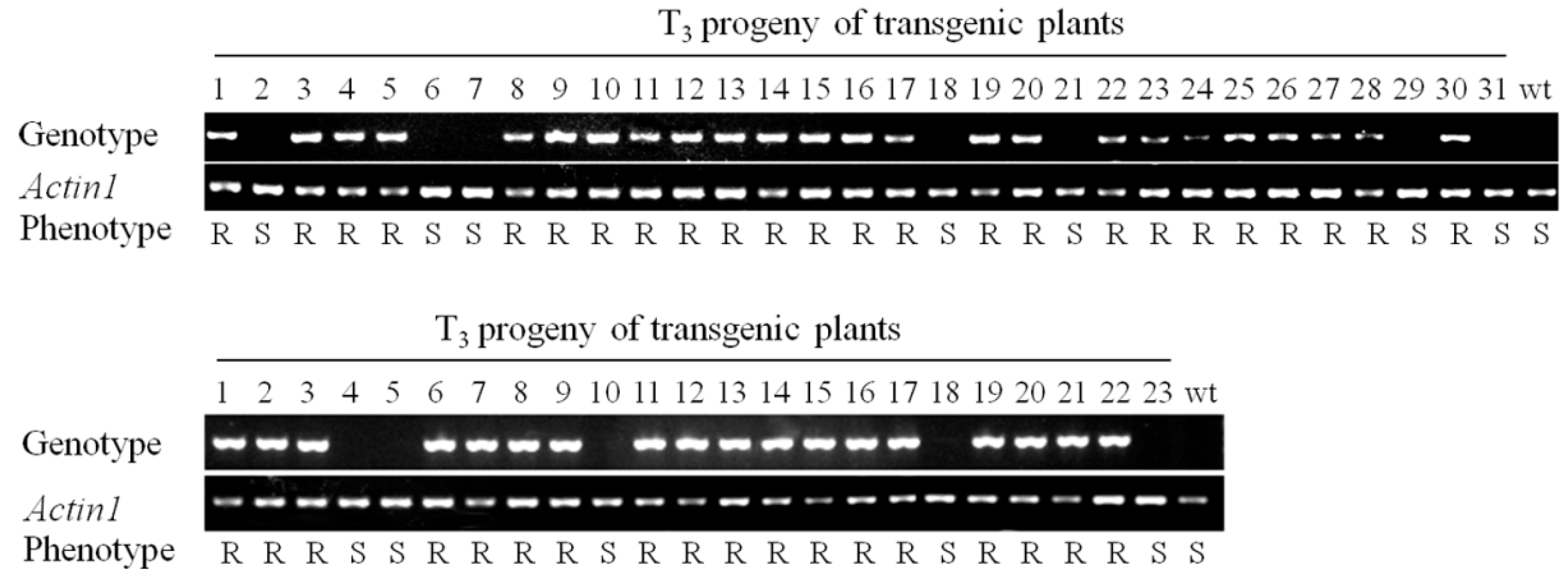
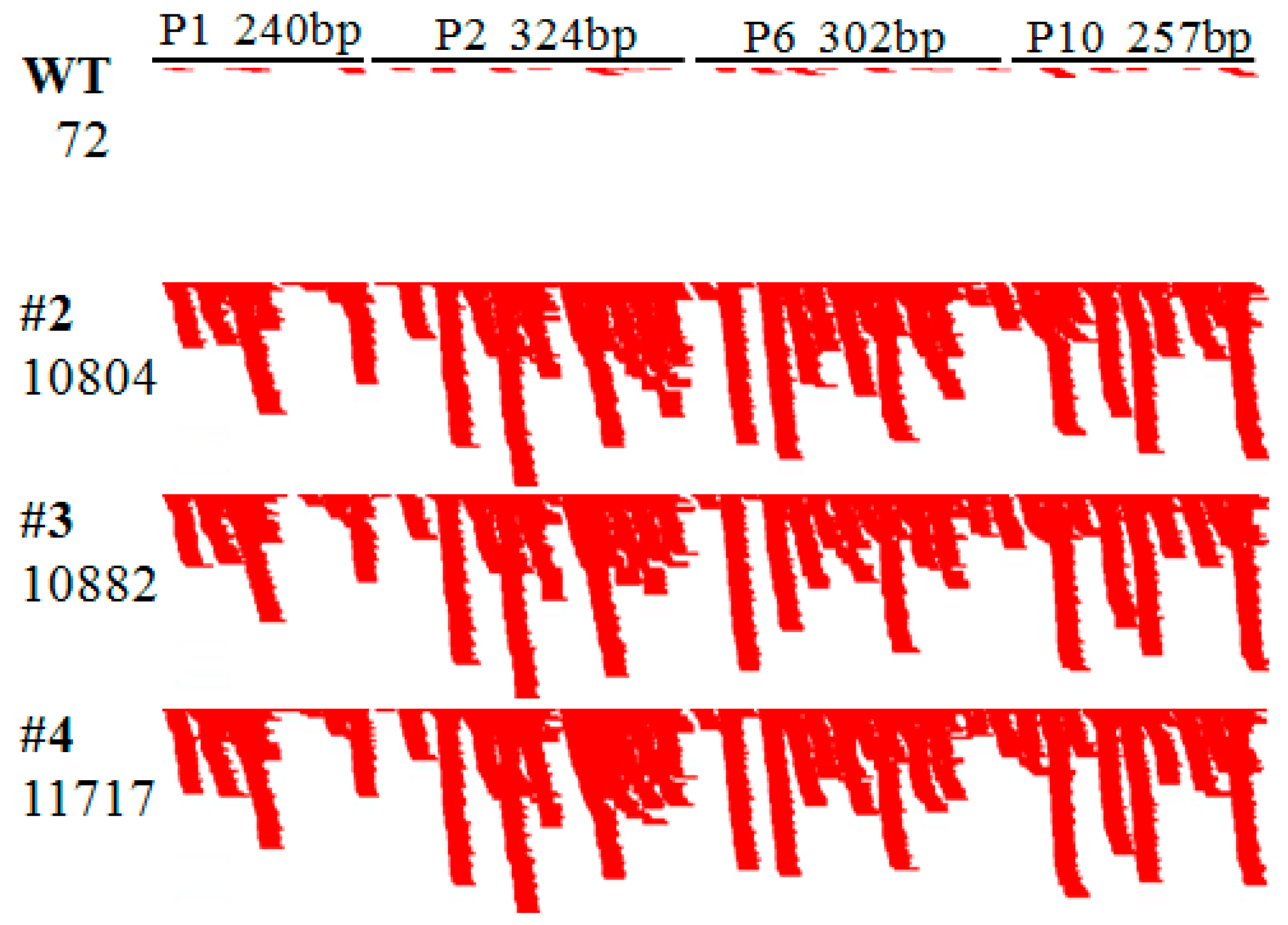
2.3. RBSDV Resistance Phenotype Is Linked to the hpRNA Transgene
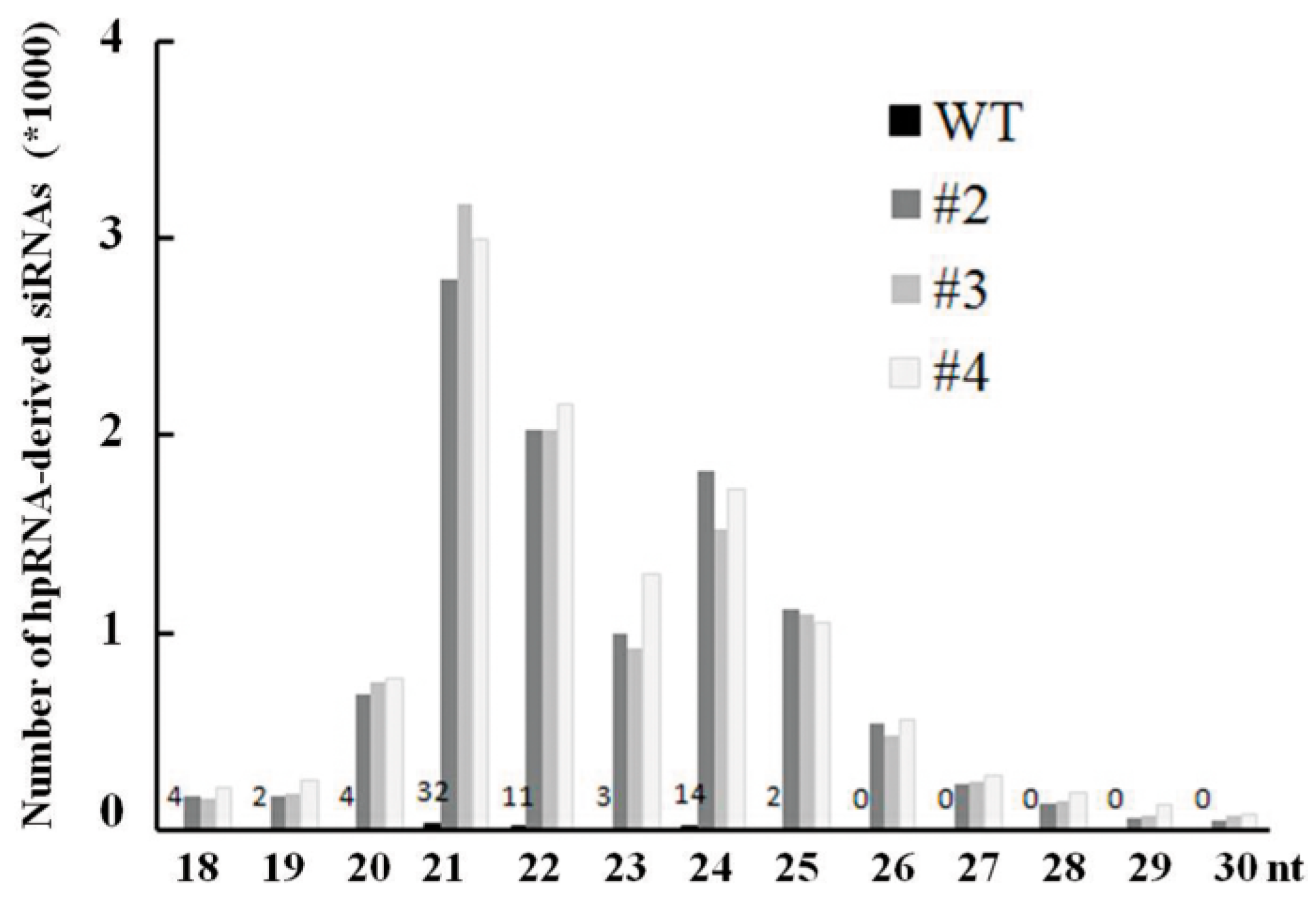
2.4. Abundant hpRNA-Derived siRNAs Are Detected in Transgenic Rice Using Small RNA Deep Sequencing
3. Discussion
4. Materials and Methods
4.1. Construction and Transformation of RNAi Vector
4.2. Plant Materials and Growth Conditions
4.3. Analysis of RBSDV Resistance
4.3.1. Disease Assay under the Field RBSDV Nursery Conditions
4.3.2. Disease Assay Using the Artificial Inoculation Method
4.4. RT-PCR
4.5. Deep Sequencing Analysis of hpRNA-Derived siRNA
5. Conclusion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fang, S.; Yu, J.; Feng, J.; Han, C.; Li, D.; Liu, Y. Identification of rice black-streaked dwarf fijivirus in maize with rough dwarf disease in China. Arch. Virol. 2001, 146, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, J.; Lei, J.; Adams, M.J. Sequence analysis shows that a dwarfing disease on rice, wheat and maize in China is caused by rice black-streaked dwarf virus. Eur. J. Plant Pathol. 2001, 107, 563–567. [Google Scholar] [CrossRef]
- Bai, F.W.; Yan, J.; Qu, Z.C.; Zhang, H.W.; Xu, J.; Ye, M.M.; Shen, D.L. Phylogenetic analysis reveals that a dwarfing disease on different cereal crops in China is due to rice black streaked dwarf virus (RBSDV). Virus Genes 2002, 25, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Ishii, M.; Yoshimura, S. Epidemiological studies on rice black-streaked dwarf virus in Kanto-Tosan district in Japan. J. Cent. Agric. Exp. Stn. 1973, 17, 61–121. [Google Scholar]
- Li, C.; Song, J.; Jiang, L. Research progress on the maize rough dwarf virus disease. Plant Prot. 1999, 25, 34–37. [Google Scholar]
- Chen, S.; Zhang, Q. Advance in researches on rice black-streaked dwarf disease and maize rough dwarf disease in China. Acta Phytophys. Sin. 2004, 32, 97–103. (In Chinese) [Google Scholar]
- Sun, F.; Xu, Q.F.; Chen, Z.B.; Fan, Y.J.; Zhou, Y.J. Advances in rice black-streaked dwarf disease in China. Jiangsu J. Agric. Sci. 2013, 29, 195–201. (In Chinese) [Google Scholar]
- Wang, Z.H.; Fang, S.G.; Xu, J.L.; Sun, L.Y.; Li, D.W.; Yu, J.L. Sequence analysis of the complete genome of rice black-streaked dwarf virus isolated from maize with rough dwarf disease. Virus Genes 2003, 27, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Chen, J.P.; Adams, M.J. Molecular characterization of segments 1 to 6 of rice black-streaked dwarf virus from China provides the complete genome. Arch. Virol. 2001, 146, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Azuhata, F.; Uyeda, I.; Kimura, I.; Shikata, E. Close similarity between genome structures of rice black-streaked dwarf and maize rough dwarf viruses. J. Gen. Virol. 1993, 74, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Isogai, M.; Uyeda, I.; Lee, B.C. Detection and assignment of proteins encoded by rice black streaked dwarf fijivirus S7, S8, S9 and S10. J. Gen. Virol. 1998, 79, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Firth, A.E.; Atkins, J.F. Analysis of the coding potential of the partially overlapping 3′ ORF in segment 5 of the plant fijiviruses. Virol. J. 2009, 6. [Google Scholar] [CrossRef] [PubMed]
- Supyani, S.; Hillman, B.I.; Suzuki, N. Baculovirus expression of the 11 mycoreovirus-1 genome segments and identification of the guanylyltransferase-encoding segment. J. Gen. Virol. 2007, 88, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Fauquet, C.M.; Mayo, M.A.; Maniloff, J.; Desselberger, U.; Ball, L.A. Virus Taxonomy: Eighth Report of the International Committee on Tax-Onomy of Viruses; Elsevier Academic Press: San Diego, CA, USA, 2005; pp. 534–542. [Google Scholar]
- Liu, H.; Wei, C.; Zhong, Y.; Li, Y. Rice black-streaked dwarf virus outer capsid protein P10 has self-interactions and forms oligomeric complexes in solution. Virus Res. 2007, 127, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, J.; Mao, X.; Wang, W.; Cheng, Z.; Zhou, Y.; Zhou, X.; Tao, X. Viroplasm protein P9-1 of Rice black-streaked dwarf virus preferentially binds to single-stranded RNA in its octamer form, and the central interior structure formed by this octamer constitutes the major RNA binding site. J. Virol. 2013, 87, 12885–12899. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xue, J.; Zhang, H.M.; Yang, J.; Xie, L.; Chen, J.P. Characterization of homologous and heterologous interactions between viroplasm proteins P6 and P9-1 of the fijivirus southern rice black-streaked dwarf virus. Arch. Virol. 2015, 160, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Z.; Wang, X.; Li, D.; Han, C.; Zhai, Y.; Yu, J. Two virus-encoded RNA silencing suppressors, P14 of beet necrotic yellow vein virus and S6 of rice black streak dwarf virus. Chin. Sci. Bull. 2005, 50, 305–310. [Google Scholar]
- Wang, Q.; Tao, T.; Zhang, Y.; Wu, W.; Li, D.; Yu, J.; Han, C. Rice black-streaked dwarf virus P6 self-interacts to form punctate, viroplasm-like structures in the cytoplasm and recruits viroplasm-associated protein P9-1. Virol. J. 2011, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, Y.; Liu, L.; Lou, Z.; Zhang, H.; Miao, H.; Hu, X.; Pang, Y.; Qiu, B. Rice black streaked dwarf virus P9-1, an α-helical protein, self-interacts and forms viroplasms in vivo. J. Gen. Virol. 2008, 89, 1770–1776. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.B.; Aragão, F.J. RNAi-mediated resistance to viruses in genetically engineered plants. Methods Mol. Biol. 2015, 1287, 81–92. [Google Scholar] [PubMed]
- Nicaise, V. Crop immunity against viruses: Outcomes and future challenges. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Sasaya, T.; Nakazono-Nagaoka, E.; Saika, H.; Aoki, H.; Hiraguri, A.; Netsu, O.; Uehara-Ichiki, T.; Onuki, M.; Toki, S.; Saito, K.; et al. Transgenic strategies to confer resistance against viruses in rice plants. Front Microbiol. 2014, 4, 409. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.; Dineshkumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The product of the tobacco mosaic virus resistance gene N similarity to toll and the interleukin-1 receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Ueda, H.; Yamaguchi, Y.; Sano, H. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 2006, 61, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Lanfermeijer, F.C.; Dijkhuis, J.; Sturre, M.J.; Haan, P.D.; Hille, J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol. Biol. 2003, 52, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Brommonschenkel, S.H.; Frary, A.; Frary, A.; Tanksley, S.D. The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol. Plant Microbe Interact. 2000, 13, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Y.; He, J.; Zheng, X.; Hu, J.; Liu, Y.; Dai, H.; Zhang, Y.; Wang, B.; Wu, W.; et al. STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat. Commun. 2014, 5, 4768. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Lee, J.H.; Park, S.K.; Hwang, U.H.; Cho, J.H.; Kwak, D.Y.; Youn, Y.N.; Yeo, U.S.; Song, Y.C.; Nam, J.; et al. Fine mapping and identification of candidate rice genes associated with qSTV11SG, a major QTL for rice stripe disease resistance. Theor. Appl. Genet. 2012, 125, 1033–1046. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakazono-Nagaoka, E.; Akita, F.; Uehara-Ichiki, T.; Omura, T.; Sasaya, T. Immunity to rice black streaked dwarf virus, a plant reovirus, can be achieved in rice plants by RNA silencing against the gene for the viroplasm component protein. Virus Res. 2011, 160, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.B.; Masuta, C.; Smith, N.A.; Shimura, H. RNA silencing and plant viral diseases. Mol. Plant Microbe Interact. 2012, 25, 1275–1285. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 2005, 6, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.F.; Matthew, L.; Smith, N.A.; Curtin, S.J.; Dedic-Hagan, J.; Ellacott, G.A.; Watson, J.M.; Wang, M.B.; Brosnan, C.; Carroll, B.J.; et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006, 7, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakazono-Nagaoka, E.; Uehara-Ichiki, T.; Sasaya, T.; Omura, T. Targeting specific genes for RNA interference is crucial to the development of strong resistance to rice stripe virus. Plant Biotechnol. J. 2011, 9, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Wu, G.; Deng, W.; Pu, Y.; Wei, C.; Li, Y. Interaction of rice dwarf virus outer capsid P8 protein with rice glycolate oxidase mediates relocalization of P8. FEBS Lett. 2007, 581, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Qian, D.; Wei, C.H.; Ye, G.; Zhang, Z.; Wu, Z.; Xie, L.; Li, Y. Movement protein Pns6 of rice dwarf phytoreovirus has both ATPase and RNA binding activities. PLoS ONE 2011, 6, e24986. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Yoshii, M.; Wei, T.; Hirochika, H.; Omura, T. Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of Rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol. J. 2009, 7, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.B.; Rezaian, A.; Watson, J.M.; Waterhouse, P.M.; Metzlaff, M.; Teixeira da Silva, J.A. Understanding and exploiting RNA silencing-mediated antiviral defense in plants. In Floriculture, Ornamental and Plant Biotechnology; Teixeira da Silva, J., Ed.; Global Science Books: London, UK, 2006; Volume III. [Google Scholar]
- Zhou, T.; Du, L.L.; Fan, Y.J.; Zhou, Y.J. Development of a RT-LAMP assay for rapid detection of rice black-streaked dwarf virus. Sci. Agric. Sin. 2012, 45, 1285–1292. (In Chinese) [Google Scholar]
- Wang, B.X.; Hu, J.L.; Sun, Z.G.; Song, Z.Q.; Lu, B.G.; Zhou, Z.L.; Fan, J.W.; Qin, D.R.; Liu, Y.Q.; Jiang, L.; et al. An evaluation system for rice black-streaked dwarf virus disease and screening for resistant rice germplasm. Acta Agron. Sin. 2014, 40, 1521–1530. (In Chinese) [Google Scholar] [CrossRef]
- Zhou, T.; Wang, Y.; Wu, L.; Fan, Y.; Zhou, Y. Method of artificial inoculation identification of rice cultivar resistance to rice black-streaked dwarf. Acta Phytophylacica Sin. 2011, 4, 301–305. (In Chinese) [Google Scholar]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson1, S.; Plaetinck, G.; Munyikwa, T.; Pleau1, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.H.; Park, S.; Zhai, J.; Gurazada, S.G.; de Paoli, E.; Meyers, B.C.; Green, P.J. Massive analysis of rice small RNAs: Mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 2011, 23, 4185–4207. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Fahlgren, N.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Carrington, J.C. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007, 5, e57. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Honda, T.; Suzuki, Y.; Kobayashi, Y.; Daito, T.; Oshida, T.; Ikuta, K.; Jern, P.; Gojobori, T.; Coffin, J.M.; et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 2010, 463, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Kondo, H.; Tani, A.; Saisho, D.; Sakamoto, W.; and Kanematsu, S.; Suzuki, N. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 2011, 7, e1002146. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ogamino, T.; Hiraguri, A.; Nakazono-Nagaoka, E.; Uehara-Ichiki, T.; Nakajima, M.; Akutsu, K.; Omura, T.; Sasaya, T. Strong resistance against Rice grassy stunt virus is induced in transgenic rice plants expressing double-stranded RNA of the viral genes for nucleocapsid or movement proteins as targets for RNA interference. Phytopathology 2013, 103, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.X.; Au, P.C.K.; Shi, B.J.; Smith, N.A.; Dennis, E.S.; Guo, H.S.; Zhou, C.Y.; Wang, M.B. Satellite RNAs interfere with the function of viral RNA silencing suppressors. Front. Plant Sci. 2015, 6, 281. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, J.; Luo, Y.; Xu, T.; Zhang, Q.; Zhang, L.; Xu, M.; Wan, J.; Wang, M.B.; Zhang, C.; et al. Construction of a genome wide RNAi mutant library in rice. Plant Biotechnol. J. 2013, 11, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Yang, J.; Zhong, W.; Zhai, H.; Zhu, L.; Fan, F.; Ali, A.J.; Yang, J.; Wang, J.; Zhu, J.; et al. Novel loci for field resistance to black-streaked dwarf and stripe viruses identified in a set of reciprocal introgression lines of rice (Oryza sativa L.). Mol. Breed. 2012, 29, 925–938. [Google Scholar] [CrossRef]
- Liu, Q.; Thorland, E.C.; Heit, J.A.; Sommer, S.S. Overlapping PCR for bidirectional PCR amplification of specific alleles: A rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes. Genome Res. 1997, 7, 389–398. [Google Scholar] [PubMed]
- Smith, N.A.; Surinder, P.S.; Wang, M.B.; Stoutjesdijk, P.A.; Green, A.G.; Waterhouse, P.M. Gene expression: Total silencing by intron-spliced hairpin RNAs. Nature 2000, 407, 319–320. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, Z.; Matthews, P.R.; Upadhyaya, N.M.; Waterhouse, P.M. Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Hortic. 1998, 461, 401–407. [Google Scholar] [CrossRef]
- Shao, M.; Wang, J.; Dean, R.A.; Lin, Y.; Gao, X.; Hu, S. Expression of a harpin-encoding gene in rice confers durable nonspecific resistance to Magnaporthe grisea. Plant Biotechnol. J. 2008, 6, 73–81. [Google Scholar] [CrossRef] [PubMed]
| Rice Materials | Disease Response of the Tested Plants | ||||
|---|---|---|---|---|---|
| N a | S b | R c | Incidence Rate (%) | ||
| Transgenic lines | #1 | 39 | 5 d | 34 | 12.82 |
| #2 | 37 | 0 | 37 | 0.00 | |
| #3 | 38 | 2 d | 36 | 5.26 | |
| #4 | 36 | 1 d | 35 | 2.78 | |
| #5 | 35 | 4 d | 31 | 11.43 | |
| Wild-type | 40 | 40 | 0 | 100.00 | |
| Huaidao 5 e | 38 | 38 | 0 | 100.00 | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Li, W.; Zhu, J.; Fan, F.; Wang, J.; Zhong, W.; Wang, M.-B.; Liu, Q.; Zhu, Q.-H.; Zhou, T.; et al. Hairpin RNA Targeting Multiple Viral Genes Confers Strong Resistance to Rice Black-Streaked Dwarf Virus. Int. J. Mol. Sci. 2016, 17, 705. https://doi.org/10.3390/ijms17050705
Wang F, Li W, Zhu J, Fan F, Wang J, Zhong W, Wang M-B, Liu Q, Zhu Q-H, Zhou T, et al. Hairpin RNA Targeting Multiple Viral Genes Confers Strong Resistance to Rice Black-Streaked Dwarf Virus. International Journal of Molecular Sciences. 2016; 17(5):705. https://doi.org/10.3390/ijms17050705
Chicago/Turabian StyleWang, Fangquan, Wenqi Li, Jinyan Zhu, Fangjun Fan, Jun Wang, Weigong Zhong, Ming-Bo Wang, Qing Liu, Qian-Hao Zhu, Tong Zhou, and et al. 2016. "Hairpin RNA Targeting Multiple Viral Genes Confers Strong Resistance to Rice Black-Streaked Dwarf Virus" International Journal of Molecular Sciences 17, no. 5: 705. https://doi.org/10.3390/ijms17050705