Gene Mutation Profiles in Primary Diffuse Large B Cell Lymphoma of Central Nervous System: Next Generation Sequencing Analyses
Abstract
:1. Introduction
2. Results
2.1. Survival Data
2.2. Mutational Profile of Primary DLBCL CNS Revealed by Targeted Next Generation Sequencing
3. Discussion
4. Experimental Sections
4.1. Patients
4.2. TruSeq Amplicon Cancer Panel Library Preparation and Sequencing
4.3. Bioinformatics Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hoang-Xuan, K.; Bessell, E.; Bromberg, J.; Hottinger, A.F.; Preusser, M.; Ruda, R.; Schlegel, U.; Siegal, T.; Soussain, C.; Abacioglu, U.; et al. Diagnosis and treatment of primary CNS lymphoma in immunocompetent patients: Guidelines from the European Association for Neuro-Oncology. Lancet Oncol. 2015, 16, e322–e332. [Google Scholar] [CrossRef]
- Montesinos-Rongen, M.; Schmitz, R.; Brunn, A.; Gesk, S.; Richter, J.; Hong, K.; Wiestler, O.D.; Siebert, R.; Kuppers, R.; Deckert, M. Mutations of CARD11 but not TNFAIP3 may activate the NF-κB pathway in primary CNS lymphoma. Acta Neuropathol. 2010, 120, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Ponzoni, M.; Issa, S.; Batchelor, T.T.; Rubenstein, J.L. Beyond high-dose methotrexate and brain radiotherapy: Novel targets and agents for primary CNS lymphoma. Ann. Oncol. 2014, 25, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Sung, C.O.; Kim, S.C.; Karnan, S.; Karube, K.; Shin, H.J.; Nam, D.H.; Suh, Y.L.; Kim, S.H.; Kim, J.Y.; Kim, S.J.; Kim, W.S.; et al. Genomic profiling combined with gene expression profiling in primary central nervous system lymphoma. Blood 2011, 117, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Tun, H.W.; Personett, D.; Baskerville, K.A.; Menke, D.M.; Jaeckle, K.A.; Kreinest, P.; Edenfield, B.; Zubair, A.C.; O’Neill, B.P.; Lai, W.R.; et al. Pathway analysis of primary central nervous system lymphoma. Blood 2008, 111, 3200–3210. [Google Scholar] [CrossRef] [PubMed]
- Bhagavathi, S.; Sharathkumar, A.; Hunter, S.; Sung, L.; Kanhere, R.; Venturina, M.D.; Wilson, J.D. Activated B-cell immunophenotype might be associated with poor prognosis of primary central nervous system lymphomas. Clin. Neuropathol. 2008, 27, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Aguilar, A.; Idbaih, A.; Boisselier, B.; Habbita, N.; Rossetto, M.; Laurenge, A.; Bruno, A.; Jouvet, A.; Polivka, M.; Adam, C.; et al. Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin. Cancer Res. 2012, 18, 5203–5211. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Boisselier, B.; Labreche, K.; Marie, Y.; Polivka, M.; Jouvet, A.; Adam, C.; Figarella-Branger, D.; Miquel, C.; Eimer, S.; et al. Mutational analysis of primary central nervous system lymphoma. Oncotarget 2014, 5, 5065–5075. [Google Scholar] [CrossRef] [PubMed]
- Luthra, R.; Patel, K.P.; Reddy, N.G.; Haghshenas, V.; Routbort, M.J.; Harmon, M.A.; Barkoh, B.A.; Kanagal-Shamanna, R.; Ravandi, F.; Cortes, J.E.; et al. Next-generation sequencing-based multigene mutational screening for acute myeloid leukemia using MiSeq: Applicability for diagnostics and disease monitoring. Haematologica 2014, 99, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Vater, I.; Montesinos-Rongen, M.; Schlesner, M.; Haake, A.; Purschke, F.; Sprute, R.; Mettenmeyer, N.; Nazzal, I.; Nagel, I.; Gutwein, J.; et al. The mutational pattern of primary lymphoma of the central nervous system determined by whole-exome sequencing. Leukemia 2014, 29, 667–685. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, B.P.; Braggio, E.; O’Neill, B.P.; van Wier, S.; Ojha, J.; McPhail, E.; Asmann, Y.; Egan, J.; Ayres da Silva, J.; Schiff, D.; et al. Genome-wide analysis uncovers recurrent alterations in primary central nervous system lymphomas (PCNSL). Neuro Oncol. 2014, 16, iii43. [Google Scholar]
- Visco, C.; Li, Y.; Xu-Monette, Z.Y.; Miranda, R.N.; Green, T.M.; Li, Y.; Tzankov, A.; Wen, W.; Liu, W.M.; Kahl, B.S.; et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: A report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia 2012, 26, 2103–2113. [Google Scholar] [PubMed]
- Braggio, E.; McPhail, E.R.; Macon, W.; Lopes, M.B.; Schiff, D.; Law, M.; Fink, S.; Sprau, D. Primary central nervous system lymphomas: A validation study of array-based comparative genomic hybridization in formalin-fixed paraffin-embedded tumor specimens. Clin. Cancer Res. 2011, 17, 4245–4253. [Google Scholar] [CrossRef] [PubMed]
- Duncavage, E.J.; Abel, H.J.; Szankasi, P.; Kelley, T.W.; Pfeifer, J.D. Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia. Mod. Pathol. 2012, 25, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; DiCarlo, J.; Satya, R.V.; Peng, Q.; Wang, Y. Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genom. 2014, 15, 244. [Google Scholar] [CrossRef] [PubMed]
- Simen, B.B.; Yin, L.; Goswami, C.P.; Davis, K.O.; Bajaj, R.; Gong, J.Z.; Peiper, S.C.; Johnson, E.S.; Wang, Z.X. Validation of a next-generation-sequencing cancer panel for use in the clinical laboratory. Arch. Pathol. Lab. Med. 2014, 139, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Sie, D.; Snijders, P.J.; Meijer, G.A.; Doeleman, M.W.; van Moorsel, M.I.; van Essen, H.F.; Eijk, P.P.; Grunberg, K.; van Grieken, N.C.; Thunnissen, E.; et al. Performance of amplicon-based next generation DNA sequencing for diagnostic gene mutation profiling in oncopathology. Cell Oncol. 2014, 37, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Kraan, W.; Horlings, H.M.; van Keimpema, M.; Schilder-Tol, E.J.; Oud, M.E.; Scheepstra, C.; Kluin, P.M.; Kersten, M.J.; Spaargaren, M.; Pals, S.T. High prevalence of oncogenic MYD88 and CD79B mutations in diffuse large B-cell lymphomas presenting at immune-privileged sites. Blood Cancer J. 2013, 3, e139. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-Rongen, M.; Brunn, A.; Bentink, S.; Basso, K.; Lim, W.K.; Klapper, W.; Schaller, C.; Reifenberger, G.; Rubenstein, J.; Wiestler, O.D.; et al. Gene expression profiling suggests primary central nervous system lymphomas to be derived from a late germinal center B cell. Leukemia 2008, 22, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-Rongen, M.; Godlewska, E.; Brunn, A.; Wiestler, O.D.; Siebert, R.; Deckert, M. Activating L265P mutations of the MYD88 gene are common in primary central nervous system lymphoma. Acta Neuropathol. 2011, 122, 791–792. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Stojanov, P.; Lawrence, M.S.; Auclair, D.; Chapuy, B.; Sougnez, C.; Cruz-Gordillo, P.; Knoechel, B.; Asmann, Y.W.; Slager, S.L.; et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 3879–3884. [Google Scholar] [CrossRef] [PubMed]
- Oren, M.; Rotter, V. Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2010, 2, a001107. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Carracedo, A.; Pandolfi, P.P. Tenets of PTEN tumor suppression. Cell 2008, 133, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.Z.; Han, D.H.; Wang, F.; Wang, Y.Q.; Zhu, Y.H.; Wu, Y.F.; Liu, N.T.; Sun, J.Y. Relationships between PTEN gene mutations and prognosis in glioma: A meta-analysis. Tumour Biol. 2014, 35, 6687–6693. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.E.; Erson-Omay, E.Z.; Serin, A.; Yin, J.; Cotney, J.; Ozduman, K.; Avsar, T.; Li, J.; Murray, P.B.; Henegariu, O.; et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 2013, 339, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Reifenberger, J.; Wolter, M.; Weber, R.G.; Megahed, M.; Ruzicka, T.; Lichter, P.; Reifenberger, G. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998, 58, 1798–1803. [Google Scholar] [PubMed]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, L.M. Primary central nervous system lymphoma: Coming or going? Blood 2009, 113, 4483–4484. [Google Scholar] [CrossRef] [PubMed]
- A quality control tool for high throughput sequence data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 2 March 2015).
- EA-utils. Available online: http://code.google.com/p/ea-utils (accessed on 2 March 2015).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- arXiv. Available online: http://arxiv.org (accessed on 2 March 2016).
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
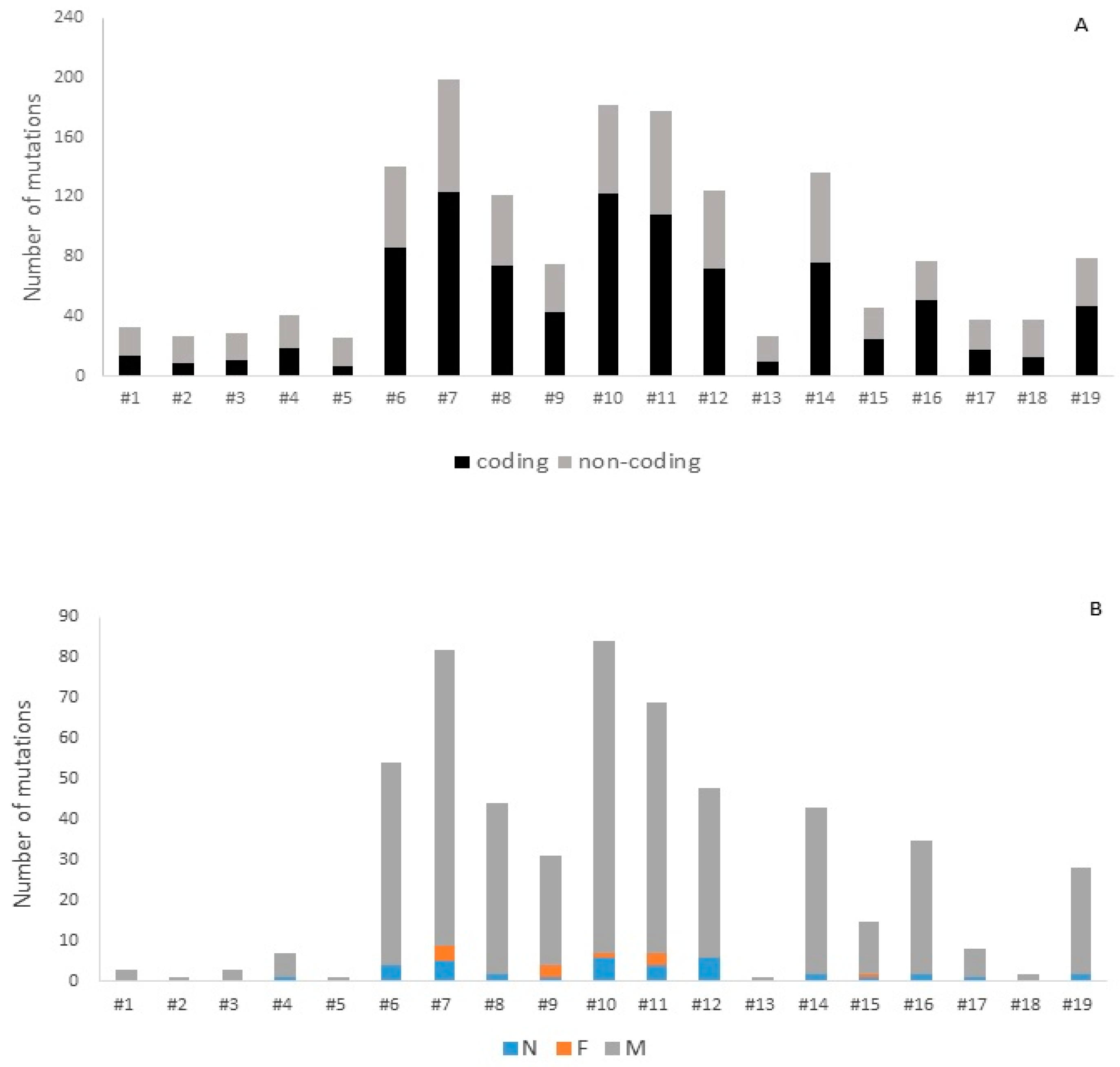
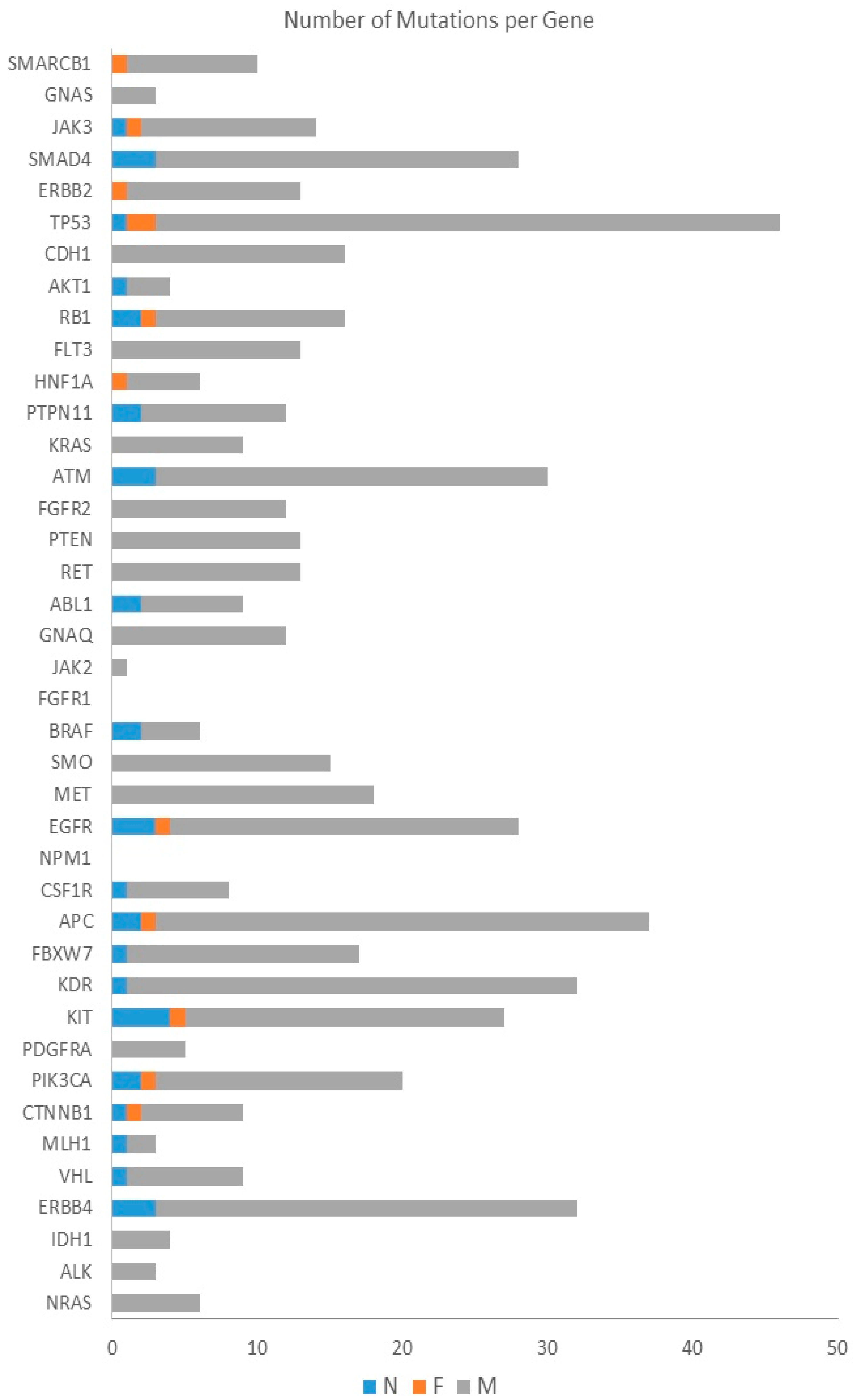
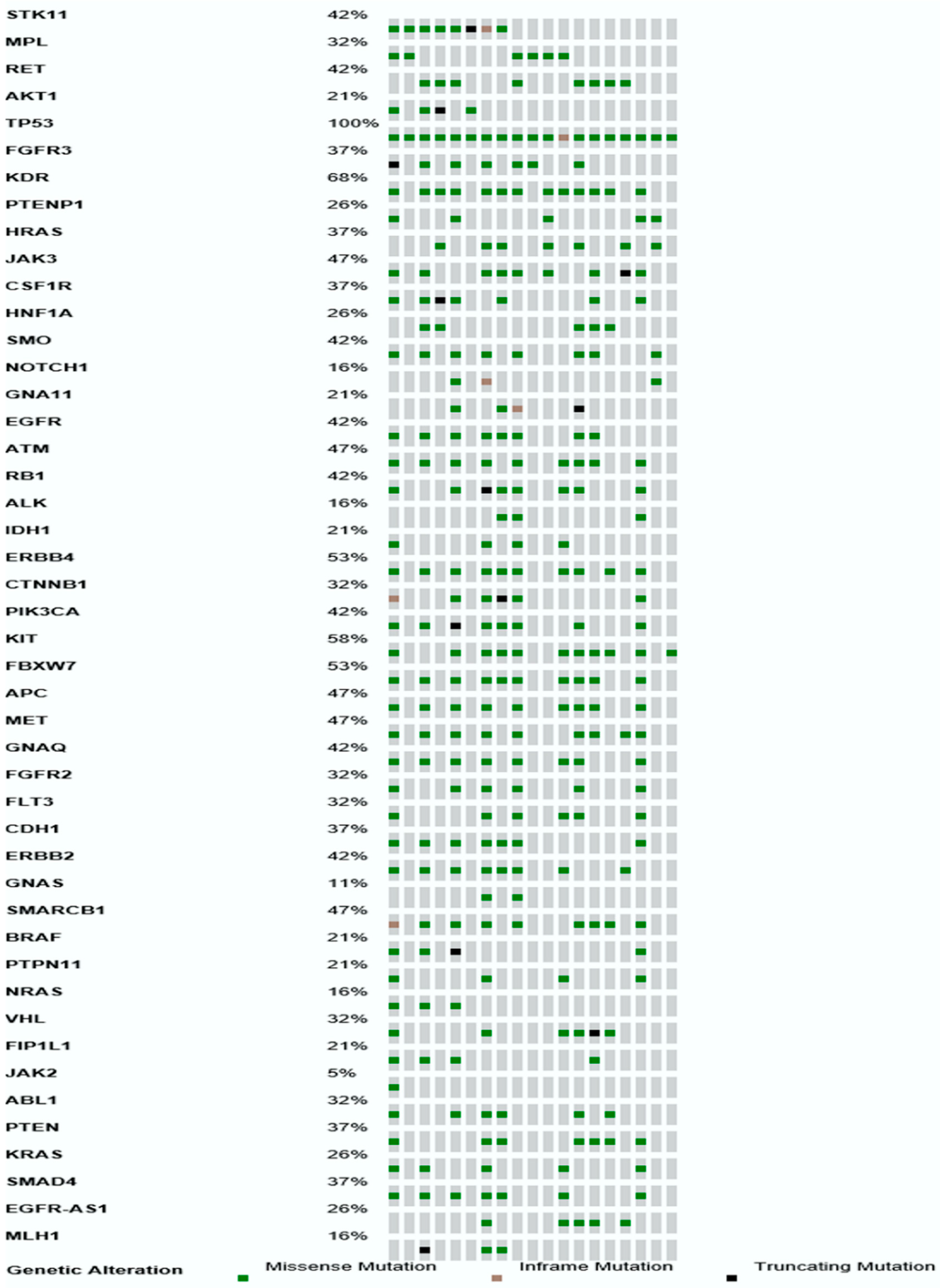
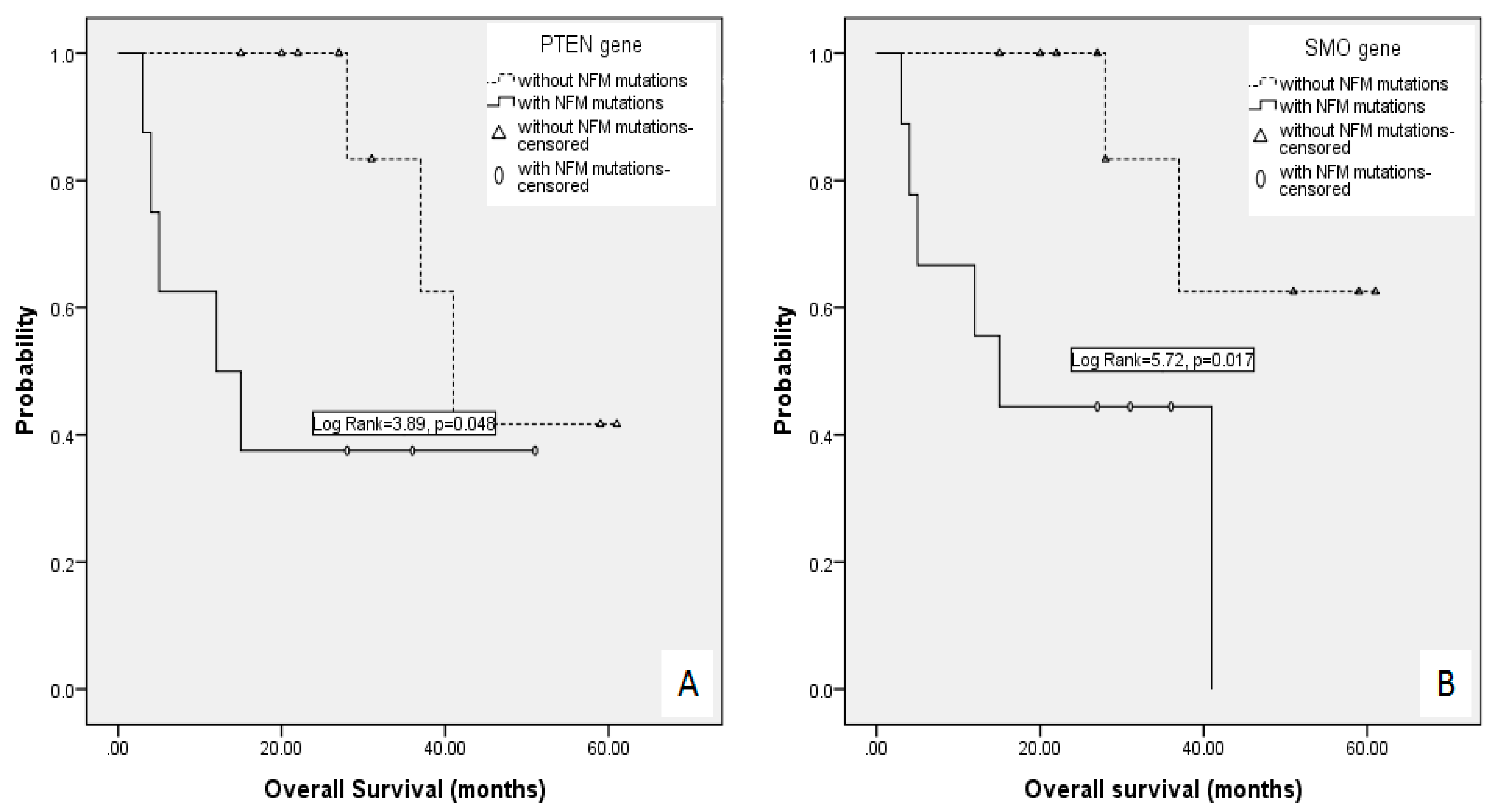
| Gene | Number of Mutations | Number of Patients |
|---|---|---|
| TP53 | 46 | 19 |
| KDR | 32 | 13 |
| KIT | 27 | 11 |
| ERBB4 | 32 | 10 |
| EGFR | 28 | 10 |
| FBXW7 | 17 | 10 |
| APC | 37 | 9 |
| ATM | 30 | 9 |
| MET | 18 | 9 |
| JAK3 | 14 | 9 |
| RET | 13 | 9 |
| PIK3CA | 20 | 8 |
| RB1 | 16 | 8 |
| ERBB2 | 13 | 8 |
| GNAQ | 12 | 8 |
| SMAD4 | 28 | 7 |
| CDH1 | 16 | 7 |
| SMO | 15 | 7 |
| PTEN | 13 | 7 |
| SMARCB1 | 10 | 7 |
| CSF1R | 8 | 7 |
| FLT3 | 13 | 6 |
| FGFR2 | 12 | 6 |
| VHL | 9 | 6 |
| CTNNB1 | 9 | 6 |
| ABL1 | 9 | 6 |
| KRAS | 9 | 5 |
| HNF1A | 6 | 5 |
| No. | Gender | Age >60 Years | ECOG PS | Tumor Localization | LDH | Visco–Young Algorithm | Treatment Approach ** | Therapy Response | Disease Relapse | Vital Status |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | No | 1 | Superficial | Normal | non-GCB | TTR | PR | Yes | Alive |
| 2 | M | No | 2 | Superficial | Normal | non-GCB | TTR | CR | Yes | Alive |
| 3 | F | Yes | 4 | Superficial | Elevated | GCB | TTR | PR | No | Alive |
| 4 | M | No | 3 | Superficial | Normal | non-GCB | PTR | CR | Yes | Dead |
| 5 | F | No | 2 | Superficial | Normal | non-GCB | TTR | CR | No | Alive |
| 6 | F | No | 3 | Deep | N/A | non-GCB | TB | CR | Yes | Dead |
| 7 | M | Yes | 4 | Superficial | N/A | non-GCB | PTR | PD | Resistant disease | Dead |
| 8 | F | No | 4 | Deep | Normal | GCB | TTR | CR | No | Alive |
| 9 | M | No | 3 | Deep | Normal | non-GCB | TB | PR | No | Alive |
| 10 | F | No | 3 | Superficial | N/A | non-GCB | PTR | PR | Yes | Dead |
| 11 | F | Yes | 1 | Superficial | Normal | non-GCB | TB | PR | No | Alive |
| 12 | F | Yes | 4 | Deep | Normal | non-GCB | PTR | PR | Yes | Dead |
| 13 | F | Yes | 3 | Deep | Elevated | non-GCB | TTR | CR | Yes | Alive |
| 14 | F | Yes | 4 | Superficial | Elevated | GCB | TTR | PD | Resistant disease | Dead |
| 15 | F | No | 4 | Superficial | Normal | non-GCB | PTR | PD | Resistant disease | Dead |
| 16 | M | No | 1 | Superficial | Elevated | non-GCB | TTR | CR | No | Alive |
| 17 | M | No | 4 | Superficial | Elevated | non-GCB | PTR | PR | Yes | Dead |
| 18 | F | No | 1 | Superficial | Normal | non-GCB | TTR | CR | Yes | Alive |
| 19 | F | No | 2 | Deep | Elevated | GCB | PTR | PR | No | Alive |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todorovic Balint, M.; Jelicic, J.; Mihaljevic, B.; Kostic, J.; Stanic, B.; Balint, B.; Pejanovic, N.; Lucic, B.; Tosic, N.; Marjanovic, I.; et al. Gene Mutation Profiles in Primary Diffuse Large B Cell Lymphoma of Central Nervous System: Next Generation Sequencing Analyses. Int. J. Mol. Sci. 2016, 17, 683. https://doi.org/10.3390/ijms17050683
Todorovic Balint M, Jelicic J, Mihaljevic B, Kostic J, Stanic B, Balint B, Pejanovic N, Lucic B, Tosic N, Marjanovic I, et al. Gene Mutation Profiles in Primary Diffuse Large B Cell Lymphoma of Central Nervous System: Next Generation Sequencing Analyses. International Journal of Molecular Sciences. 2016; 17(5):683. https://doi.org/10.3390/ijms17050683
Chicago/Turabian StyleTodorovic Balint, Milena, Jelena Jelicic, Biljana Mihaljevic, Jelena Kostic, Bojana Stanic, Bela Balint, Nadja Pejanovic, Bojana Lucic, Natasa Tosic, Irena Marjanovic, and et al. 2016. "Gene Mutation Profiles in Primary Diffuse Large B Cell Lymphoma of Central Nervous System: Next Generation Sequencing Analyses" International Journal of Molecular Sciences 17, no. 5: 683. https://doi.org/10.3390/ijms17050683






