Mycoplasma bovis MBOV_RS02825 Encodes a Secretory Nuclease Associated with Cytotoxicity
Abstract
:1. Introduction
2. Results
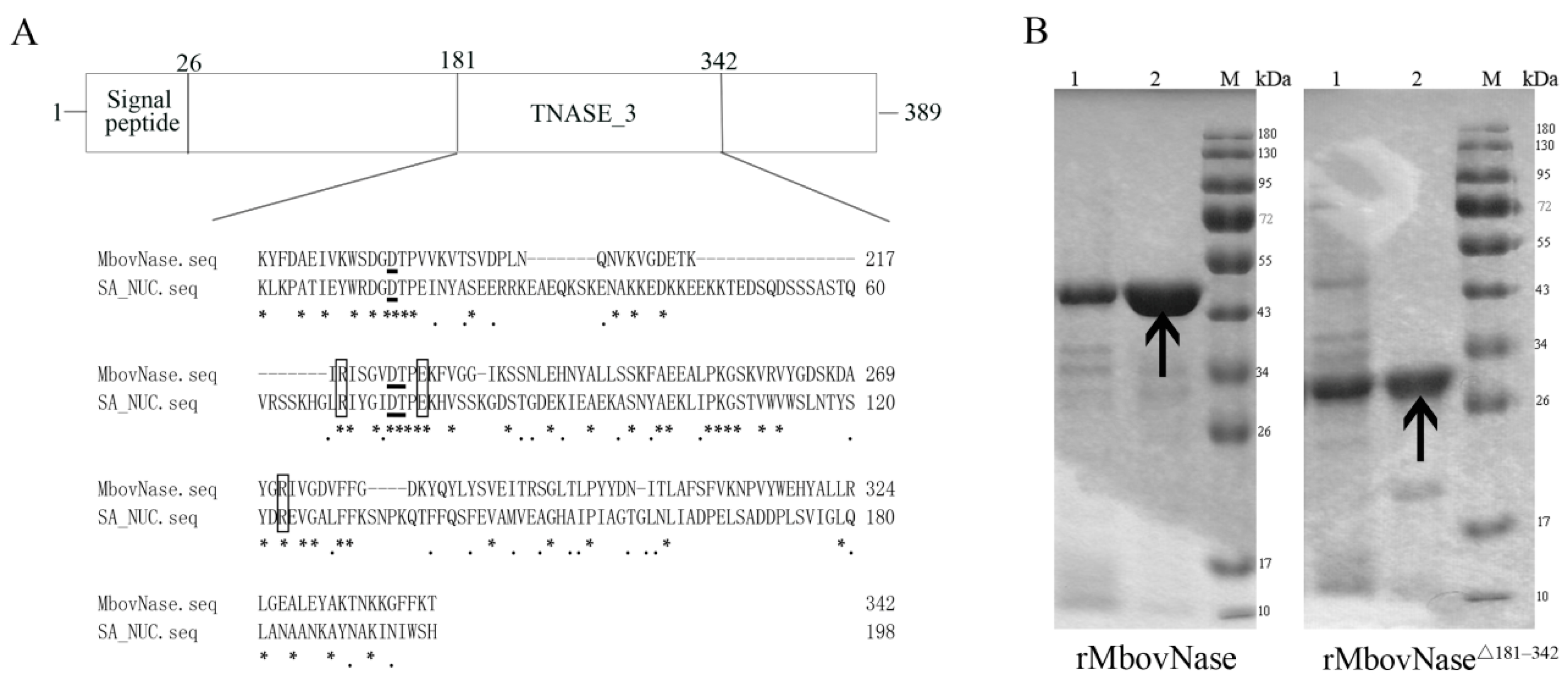
2.1. MBOV_RS02825 Sequence Has a Typical Nuclease Structure
2.2. Expression of rMbovNase in Escherichia coli
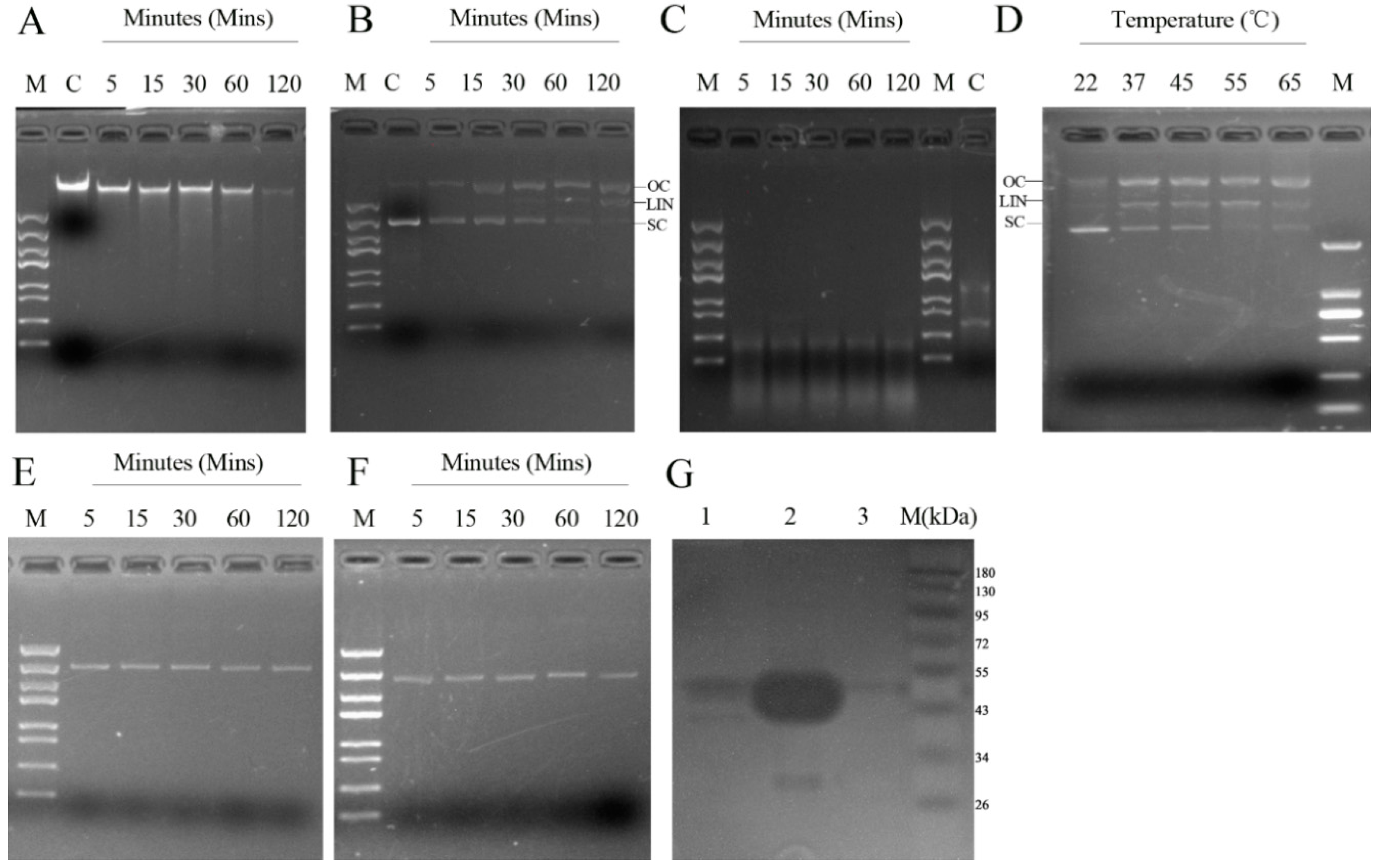
2.3. Determinants of rMbovNase Nuclease Activity
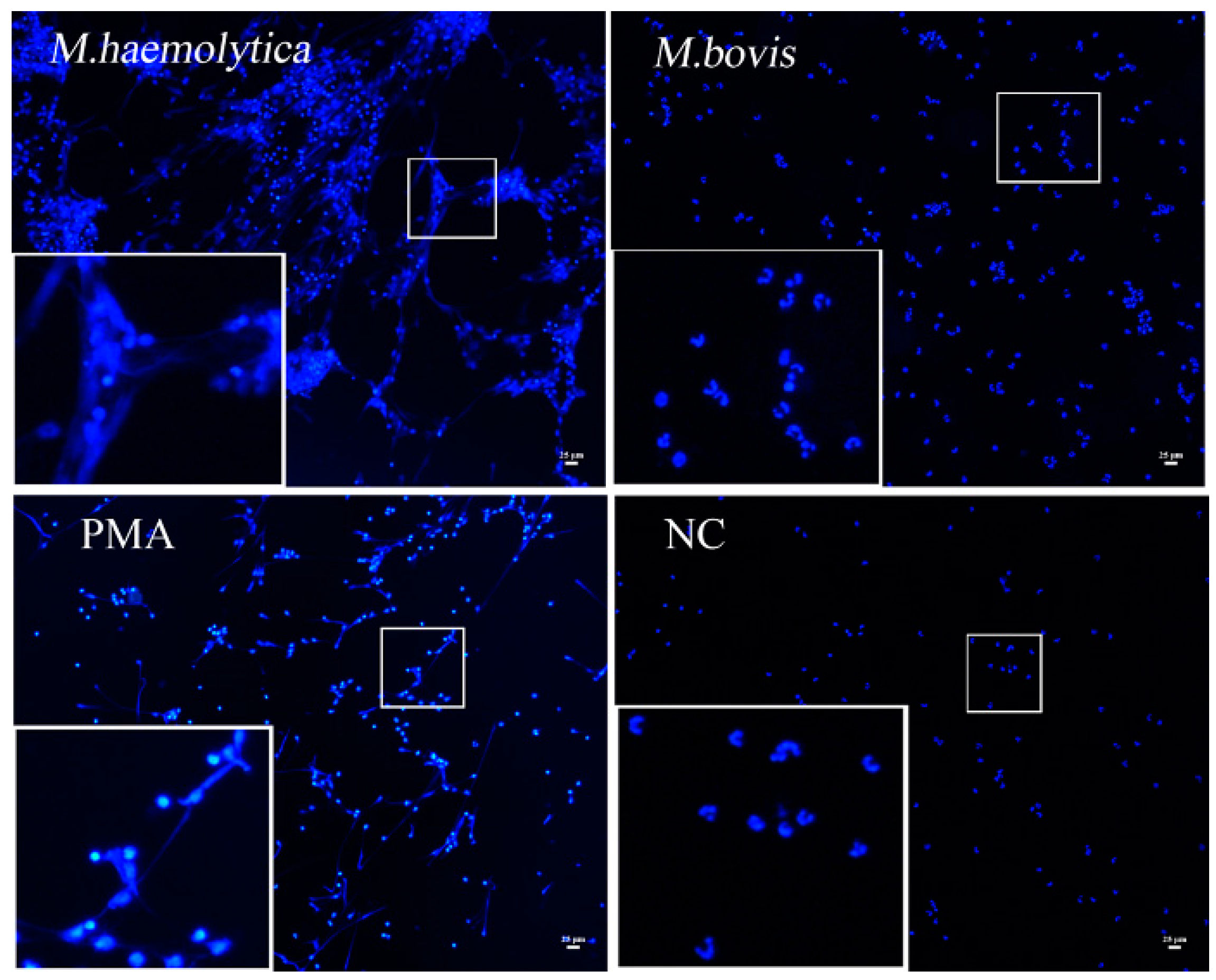
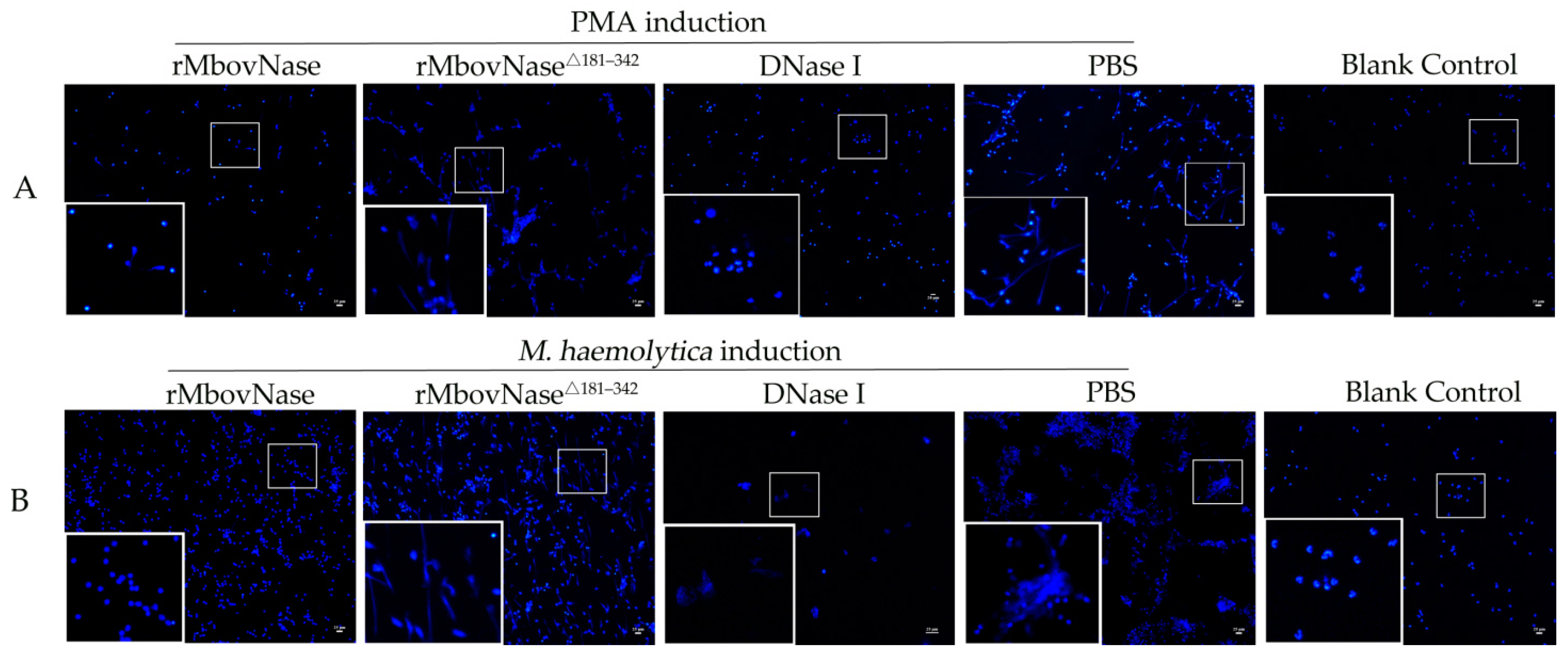
2.4. rMbovNase Breakdown of NETs
2.5. Localization of MbovNase in M. bovis Cells
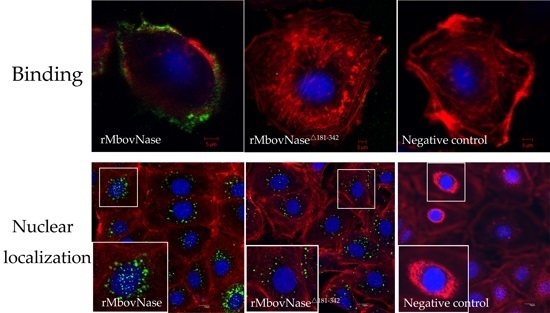
2.6. TNASE_3 Is Essential for rMbovNase Binding and Internalization in BoMac Cells
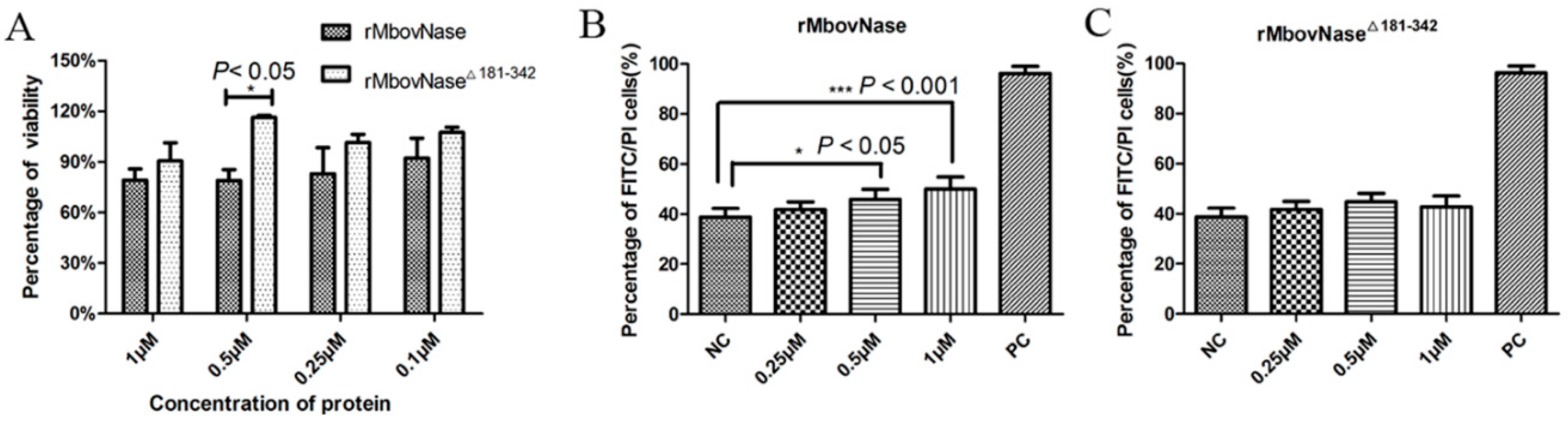
2.7. rMbovNase-Induced Cytotoxicity and Apoptosis of BoMac Cells
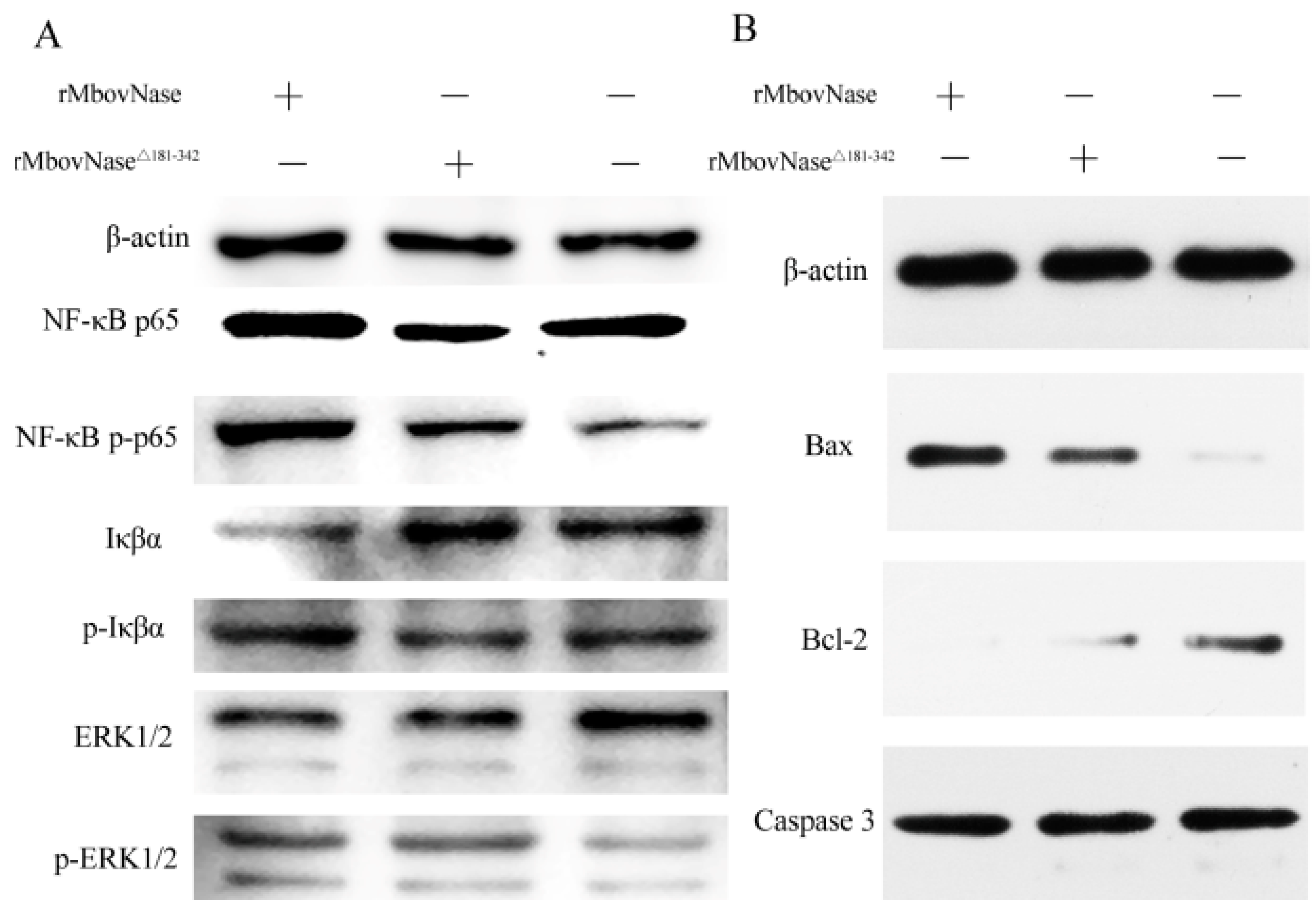
2.8. Expression of Some Signal Molecules Associated with Apoptosis
3. Discussion
3.1. rMbovNase Is a Membrane Bound and Secretory Nuclease
3.2. Binding, Internalization, and Cytotoxicity of rMbovNase
4. Materials and Methods
4.1. Ethics Statement
4.2. Bacterial Strains, Plasmids and DNA Manipulation
4.3. Cell Culture Conditions
4.4. Computer-Assisted Sequence Analysis
4.5. Cloning and Expression of MBOV_RS02825 and Generation of Antiserum
4.6. Analysis of rMbovNase Nuclease Activity
4.7. Zymogram Analysis
4.8. NETs Formation and Degradation Assay
4.9. Localization of rMbovNase by Western Blot Analysis
4.10. rMbovNase Binding and Invasion Assay
4.11. MTT Assay of Cell Viability after rMbovNase Treatment
4.12. Detection of Cell Apoptosis Induced by rMbovNase
4.13. Western Blot Assay of Molecular Expression Related to Apoptosis and the NF-κB Signal Pathway
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hale, H.H.; Helmboldt, C.F.; Plastridge, W.N.; Stula, E.F. Bovine mastitis caused by a mycoplasma species. Cornell Vet. 1962, 52, 582–591. [Google Scholar] [PubMed]
- Maunsell, F.P.; Woolums, A.R.; Francoz, D.; Rosenbusch, R.F.; Step, D.L.; Wilson, D.J.; Janzen, E.D. Mycoplasma bovis infections in cattle. J. Vet. Intern. Med. 2011, 25, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.; Li, Y.; Guo, D.; Song, N.; Hu, S.; Chen, C.; Pei, J.; Cao, P. First isolation of Mycoplasma bovis from calf lung with pneumoniae in China. Chin. J. Prev. Vet. Med. 2008, 30, 661–664. (In Chinese) [Google Scholar]
- Minion, F.C.; Jarvill-Taylor, K.J.; Billings, D.E.; Tigges, E. Membrane-associated nuclease activities in mycoplasmas. J. Bacteriol. 1993, 175, 7842–7847. [Google Scholar] [PubMed]
- Schmidt, J.A.; Browning, G.F.; Markham, P.F. Mycoplasma hyopneumoniae mhp379 is a Ca2+-dependent, sugar-nonspecific exonuclease exposed on the cell surface. J. Bacteriol. 2007, 189, 3414–3424. [Google Scholar] [CrossRef] [PubMed]
- Somarajan, S.R.; Kannan, T.R.; Baseman, J.B. Mycoplasma pneumoniae Mpn133 is a cytotoxic nuclease with a glutamic acid-, lysine- and serine-rich region essential for binding and internalization but not enzymatic activity. Cell. Microbiol. 2010, 12, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Teng, D.; Jiang, F.; Zhang, Y.; El-Ashram, S.A.; Wang, H.; Sun, Z.; He, J.; Shen, J.; Wu, W.; et al. Mycoplasma gallisepticum MGA_0676 is a membrane-associated cytotoxic nuclease with a staphylococcal nuclease region essential for nuclear translocation and apoptosis induction in chicken cells. Appl. Microbiol. Biotechnol. 2015, 99, 1859–1871. [Google Scholar] [CrossRef] [PubMed]
- Bendjennat, M.; Blanchard, A.; Loutfi, M.; Montagnier, L.; Bahraoui, E. Role of Mycoplasma penetrans endonuclease p40 as a potential pathogenic determinant. Infect. Immun. 1999, 67, 4456–4462. [Google Scholar] [PubMed]
- Li, L.; Krishnan, M.; Baseman, J.B.; Kannan, T.R. Molecular cloning, expression, and characterization of a Ca2+-dependent, membrane-associated nuclease of Mycoplasma genitalium. J. Bacteriol. 2010, 192, 4876–4884. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Tivendale, K.A.; Markham, P.F.; Browning, D.F. Disruption of the membrane nuclease gene (MBOVPG45_0215) of Mycoplasma bovis greatly reduces cellular nuclease activity. J. Bacteriol. 2015, 197, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Hynes, T.R.; Fox, R.O. The crystal structure of staphylococcal nuclease refined at 1.7 Å resolution. Proteins 1991, 10, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Berends, E.T.; Horswill, A.R.; Haste, N.M.; Monestier, M.; Nizet, V.; von Kockritz-Blickwede, M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2010, 2, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Von Kockritz-Blickwede, M.; Nizet, V. Innate immunity turned inside-out: Antimicrobial defense by phagocyte extracellular traps. J. Mol. Med. (Berl.) 2009, 87, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.È.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Rahman, O.; Cummings, S.P.; Harrington, D.J.; Sutcliffe, L.C. Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J. Microb. Biotechnol. 2008, 24, 2377–2382. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- IPP6.0. Available online: http://www.mediacy.com/index.aspx?page=IPP (accessed on 15 December 2015).
- Djordjevic, S.P.; Cordwell, S.J.; Djordjevic, M.A.; Wilton, J.; Minion, F.C. Proteolytic processing of the Mycoplasma hyopneumoniae cilium adhesion. Infect. Immun. 2004, 72, 2791–2802. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.L.; Wise, K.S. Site-specific proteolysis of the MALP-404 lipoprotein determines the release of a soluble selective lipoprotein-associated motif-containing fragment and alteration of the surface phenotype of Mycoplasma fermentans. Infect. Immun. 2002, 70, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Hopfe, M.; Hoffmann, R.; Henrich, B. P80, the HinT interacting membrane protein, is a secreted antigen of Mycoplasma hominis. BMC Microbiol. 2004, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razin, S.; Knyszynski, A.; Lifshitz, Y. Nucleases of mycoplasma. J. Gen. Microbiol. 1964, 36, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Aebi, M.; Bodmer, M.; Frey, J.; Pilo, P. Herd-specific strains of Mycoplasma bovis in outbreaks of mycoplasmal mastitis and pneumonia. Vet. Microbiol. 2012, 157, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Maunsell, F.; Brown, M.B.; Powe, J.; Ivey, J.; Woolard, M.; Love, W.; Simecka, J.W. Oral inoculation of young dairy calves with Mycoplasma bovis results in colonization of tonsils, development of otitis media and local immunity. PLoS ONE 2012, 7, e44523. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wang, G.; Lv, T.; Song, X.; Wang, T.; Xie, G.; Cao, Y.; Zhang, N.; Cao, R. Endometrial inflammation and abnormal expression of extracellular matrix proteins induced by Mycoplasma bovis in dairy cows. Theriogenology 2014, 81, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Feissner, R.F.; Skalska, J.; Gaum, W.E.; Sheu, S.S. Crosstalk signaling between mitochondrial Ca2+ and ROS. Front. Biosci. (Landmark Ed.) 2009, 14, 1197–1218. [Google Scholar] [CrossRef] [PubMed]
- Schott, C.; Cai, H.; Parker, L.; Bateman, K.G.; Caswell, J.L. Hydrogen peroxide production and free radical-mediated cell stress in Mycoplasma bovis pneumonia. J. Comp. Pathol. 2014, 150, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Cacciotto, C.; Addis, M.F.; Coradduzza, E.; Carcangiu, L.; Nuvoli, A.M.; Tore, G.; Dore, G.M.; Pagnozzi, D.; Uzzau, S.; Chessa, B.; et al. Mycoplasma agalactiae MAG_5040 is a Mg2+-dependent, sugar-nonspecific snase recognised by the host humoral response during natural infection. PLoS ONE 2013, 8, e57775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masukagami, Y.; Tivendale, K.A.; Mardani, K.; Ben-Barak, I.; Markham, P.F.; Browning, G.F. The Mycoplasma gallisepticum virulence factor lipoprotein MslA is a novel polynucleotide binding protein. J. Innate Immun. 2013, 81, 3220–3226. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.F.; Reichard, U.; Brinkmann, V.; Zychlinsky, A. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 2006, 8, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Chow, O.A.; von Köckritz-Blickwede, M.; Bright, A.T.; Hensler, M.E.; Zinkernagel, A.S.; Cogen, A.L.; Gallo, R.L.; Monestier, M.; Wang, Y.; Glass, C.K.; et al. Statins enhance formation of phagocyte extracellular traps. Cell Host Microbe. 2010, 8, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Paunel-Gorgulu, A.; Flohe, S.; Witte, I.; Schadel-Hopfner, M.; Windolf, J.; Logters, T.T. Deoxyribonuclease is a potential counter regulator of aberrant neutrophil extracellular traps formation after major trauma. Mediat. Inflamm. 2012, 149560. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.; Halverson, T.W.R.; Wilton, M.; Poon, K.K.H.; Petri, B.; Lewenza, S. DNA is an antimicrobial component of neutrophil extracellular traps. PLOS Pathog. 2015, 11, e1004593. [Google Scholar]
- Derre-Bobillot, A.; Cortes-Perez, N.G.; Yamamoto, Y.; Kharrat, P.; Couve, E.; Da Cunha, V.; Decker, P.; Boissier, M.C.; Escartin, F.; Cesselin, B.; et al. Nuclease a (GBS0661), an extracellular nuclease of Streptococcus agalactiae, attacks the neutrophil extracellular traps and is needed for full virulence. Mol. Microbiol. 2013, 89, 518–531. [Google Scholar] [CrossRef] [PubMed]
- De Buhr, N.; Neumann, A.; Jerjomiceva, N.; von Kockritz-Blickwede, M.; Baums, C.G. Streptococcus suis dnase Ssna contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiology 2014, 160, 385–395. [Google Scholar] [CrossRef] [PubMed]
- De Buhr, N.; Stehr, M.; Neumann, A.; Naim, H.Y.; Valentin-Weigand, P.; von Kockritz-Blickwede, M.; Baums, C.G. Identification of a novel DNase of streptococcus suis (EndAsuis) important for neutrophil extracellular trap degradation during exponential growth. Microbiology 2015, 161, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Gabinaitiene, A.; Siugzdaite, J.; Zilinskas, H.; Siugzda, R.; Petkevicius, S. Mycoplasma bovis and bacterial pathogens in the bovine respiratory tract. Vet. Med. Czech 2011, 56, 28–34. [Google Scholar]
- Aulik, N.A.; Hellenbrand, K.M.; Klos, H.; Czuprynski, C.J. Mannheimia haemolytica and its leukotoxin cause neutrophil extracellular trap formation by bovine neutrophils. Infect. Immun. 2010, 78, 4454–4466. [Google Scholar] [CrossRef] [PubMed]
- Burki, S.; Frey, J.; Pilo, P. Virulence, persistence and dissemination of Mycoplasma bovis. Vet. Microbiol. 2015, 179, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Adamu, J.Y.; Wawegama, N.K.; Browning, G.F.; Markham, P.F. Membrane proteins of Mycoplasma bovis and their role in pathogenesis. Res. Vet. Sci. 2013, 95, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y.; Luo, X.; Wang, X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 2001, 412, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Ueno, P.M.; Timenetsky, J.; Centonze, V.E.; Wewer, J.J.; Cagle, M.; Stein, M.A.; Krishnan, M.; Baseman, J.B. Interaction of Mycoplasma genitalium with host cells: Evidence for nuclear localization. Microbiology 2008, 154, 3033–3041. [Google Scholar] [CrossRef] [PubMed]
- Saini, A.K.; Maithal, K.; Chand, P.; Chowdhury, S.; Vohra, R.; Goyal, A.; Dubey, G.P.; Chopra, P.; Chandra, R.; Tyagi, A.K.; et al. Nuclear localization and in situ DNA damage by Mycobacterium tuberculosis nucleoside-diphosphate kinase. J. Biol. Chem. 2004, 279, 50142–50149. [Google Scholar] [CrossRef] [PubMed]
- Zurawski, D.V.; Mitsuhata, C.; Mumy, K.L.; McCormick, B.A.; Maurelli, A.T. OspF and OspC1 are Shigella flexneri type III secretion system effectors that are required for postinvasion aspects of virulence. Infect. Immun. 2006, 74, 5964–5976. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Drapkin, R.; Yalamanchili, R.; Mosialos, G.; Kieff, E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 1995, 15, 4735–4744. [Google Scholar] [CrossRef] [PubMed]
- Elliott, G.; O’Hare, P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 1997, 88, 223–233. [Google Scholar] [CrossRef]
- Wender, P.A.; Mitchell, D.J.; Pattabiraman, K.; Pelkey, E.T.; Steinman, L.; Rothbard, J.B. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: Peptoid molecular transporters. Proc. Natl. Acad. Sci. USA 2000, 97, 13003–13008. [Google Scholar] [CrossRef] [PubMed]
- Potocky, T.B.; Menon, A.K.; Gellman, S.H. Cytoplasmic and nuclear delivery of a tat-derived peptide and a β-peptide after endocytic uptake into hela cells. J. Biol. Chem. 2003, 278, 50188–50194. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, I.A.; Vaughan, A.T.; Khodarev, N.N. Mycoplasma infection can sensitize host cells to apoptosis through contribution of apoptosis-like endonuclease(s). Immunol. Cell Biol. 1998, 76, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Guo, A.; Cui, P.; Chen, Y.; Mustafa, R.; Ba, X.; Hu, C.; Bai, Z.; Chen, X.; Shi, L.; et al. Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese isolate). PLoS ONE 2012, 7, e38239. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; J, P.; Peng, Q.; Lai, J.; Liu, Y.; Tong, S.; Shao, Y.; Chen, Y.; Hu, C.; Guo, A. Study on comparative identification for cattle Mannheimia haemolytica, Pasteurella multocida type A and B strains. China Dairy Cattle. 2015, 14, 48–53. (In Chinese) [Google Scholar]
- Stabel, J.R.; Stabel, T.J. Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet. Immunol. Immunopathol. 1995, 45, 211–220. [Google Scholar] [CrossRef]
- MBOV_RS02825 sequence. Available online: http://www.ncbi.nlm.nih.gov/protein/AFM51934.1 (accessed on 1 December 2015).
- PROSITE. Available online: http://prosite.expasy.org/prosite.html (accessed on 10 Feburary 2016).
- BLASTP. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 Feburary 2016).
- TMHMM 2.0. Available online: http://www.cbs.dtu.dk/services (accessed on 10 Feburary 2016).
- Zhao, J.; Pan, S.; Lin, L.; Fu, L.; Yang, C.; Xu, Z.; Wei, Y.; Jin, M.; Zhang, A. Streptococcus suis serotype 2 strains can induce the formation of neutrophil extracellular traps and evade trapping. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Li, Y.; Wang, Y.; Zhou, Y.; Liu, Y.; Xin, J. Molecular cloning and characterization of a surface-localized adhesion protein in Mycoplasma bovis Hubei-1 strain. PLoS ONE 2013, 8, e69644. [Google Scholar] [CrossRef] [PubMed]


| Names | Primer Sequences (5′-3′) | Positions |
|---|---|---|
| MBOV_RS02825 | ||
| 580F1 | CGGGGTACCGAAAATGGCACAATTAAG | 79→97 |
| 580R1 | AGTTTTTTCTCCCAGCCATCTACTAACTC | 316←345 |
| 580F2 | GTTAGTAGATGGCTGGGAGAAAAAACTTAG | 317→347 |
| 580R2 | AATATTTAGCCTTCAATTTGTCCCAATCTATG | 437←469 |
| 580F3 | CTAAATATTTTGATGCAGAAATAGTTAAATGGAGCG | 460→496 |
| 580R3 | GGCATAATGCTCCCAGTAAACTGGATTTTT | 855←885 |
| 580F4 | AATCCAGTTTACTGGGAGCATTATGCC | 855→885 |
| 580R4 | TAAATGTTAGATTGAATCCAGTATGGC | 956←983 |
| 580F5 | GCCATACTGGATTCAATCTAACATTTA | 956→983 |
| 580R5 | AGTGTTTATCTAGCAATGTCCATTTTC | 1000←1027 |
| 580F6 | TGGAAAATGGACATTGCTAGATAAACA | 998→1025 |
| 580R6 | CGCGGATCCTTATTTATTTTTGTATGAATC | 1071←1092 |
| MBOV_RS02825ΔTNASE_3 | ||
| 580F1 | CGGGGTACCGAAAATGGCACAATTAAG | 79→97 |
| 580UR | TGGCAATGCAAATTTAGCCTTCAATTT | 530←543, 1027←1040 |
| 580DF | AAATTGAAGGCTAAATTTGCATTGCCA | 530→543, 1027→1040 |
| 580R6 | CGCGGATCCTTATTTATTTTTGTATGAATC | 1071←1092 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhao, G.; Guo, Y.; Menghwar, H.; Chen, Y.; Chen, H.; Guo, A. Mycoplasma bovis MBOV_RS02825 Encodes a Secretory Nuclease Associated with Cytotoxicity. Int. J. Mol. Sci. 2016, 17, 628. https://doi.org/10.3390/ijms17050628
Zhang H, Zhao G, Guo Y, Menghwar H, Chen Y, Chen H, Guo A. Mycoplasma bovis MBOV_RS02825 Encodes a Secretory Nuclease Associated with Cytotoxicity. International Journal of Molecular Sciences. 2016; 17(5):628. https://doi.org/10.3390/ijms17050628
Chicago/Turabian StyleZhang, Hui, Gang Zhao, Yusi Guo, Harish Menghwar, Yingyu Chen, Huanchun Chen, and Aizhen Guo. 2016. "Mycoplasma bovis MBOV_RS02825 Encodes a Secretory Nuclease Associated with Cytotoxicity" International Journal of Molecular Sciences 17, no. 5: 628. https://doi.org/10.3390/ijms17050628