Identification and Comparative Analysis of CBS Domain-Containing Proteins in Soybean (Glycine max) and the Primary Function of GmCBS21 in Enhanced Tolerance to Low Nitrogen Stress
Abstract
:1. Introduction
2. Results
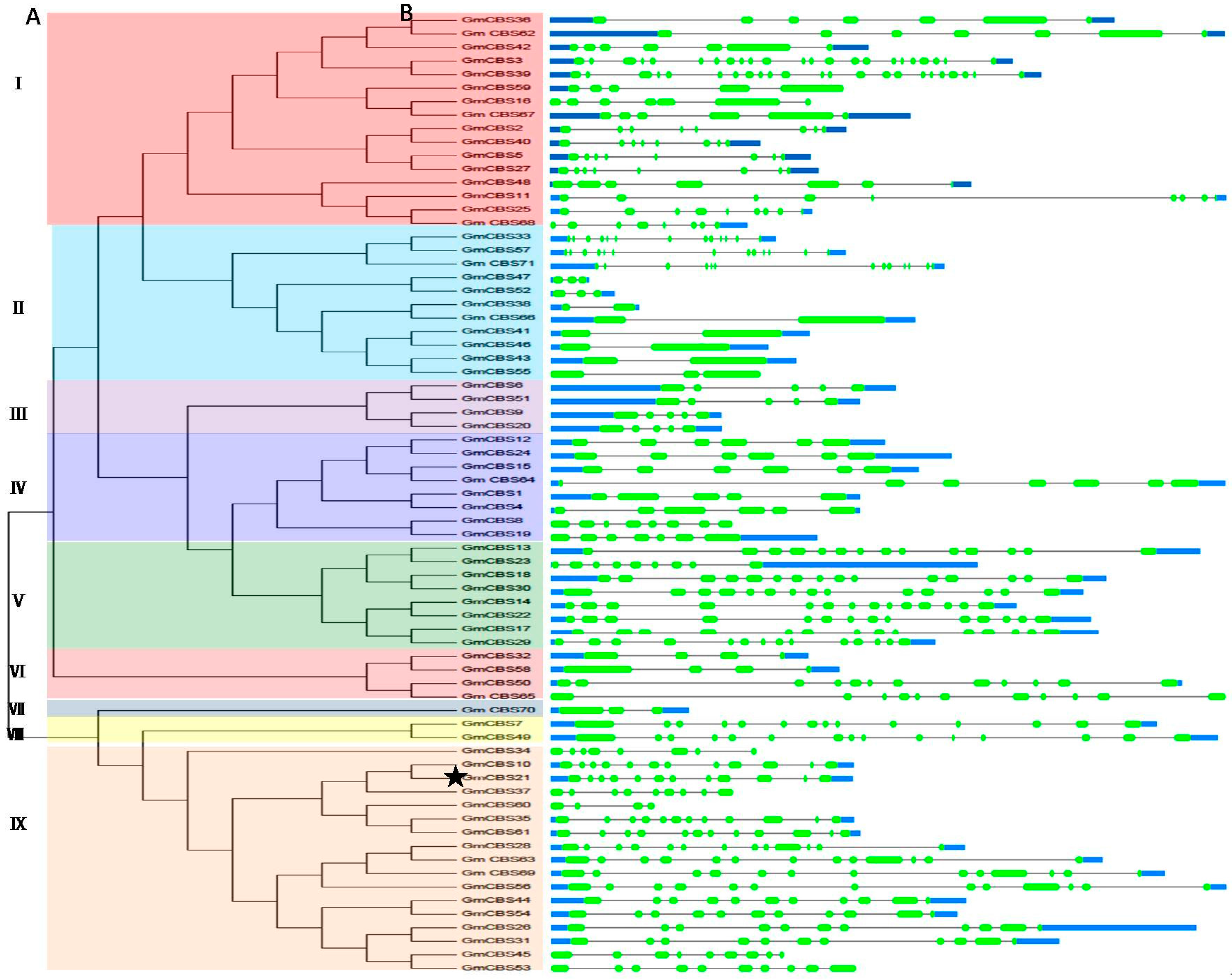
2.1. Identification, Characterization and Classification of CBS Domain-Containing Proteins in Soybean
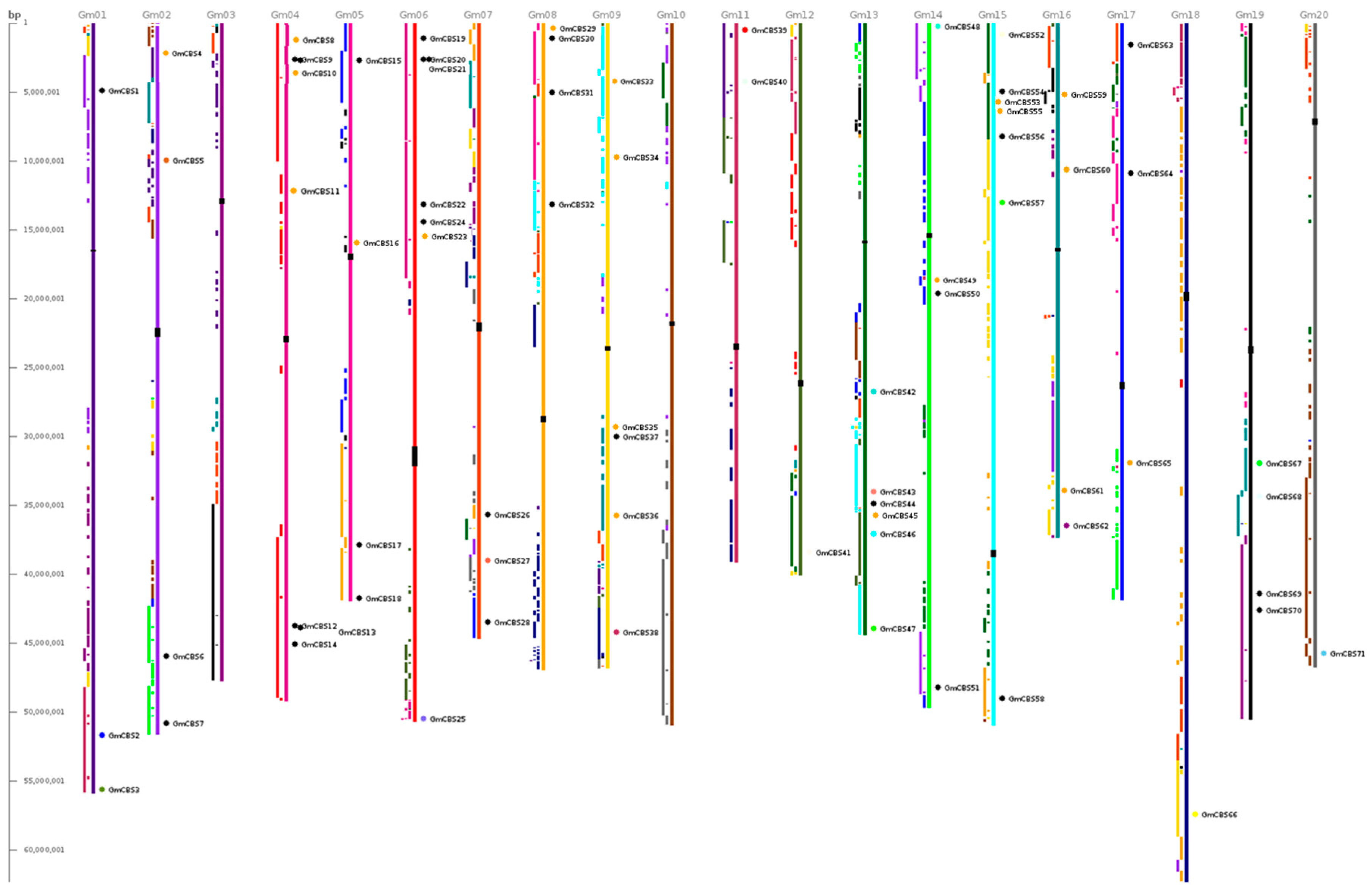
2.2. Phylogenetic Motif Recognition, Genome Distribution and Gene Structure of the CBS Superfamily Genes in Soybean
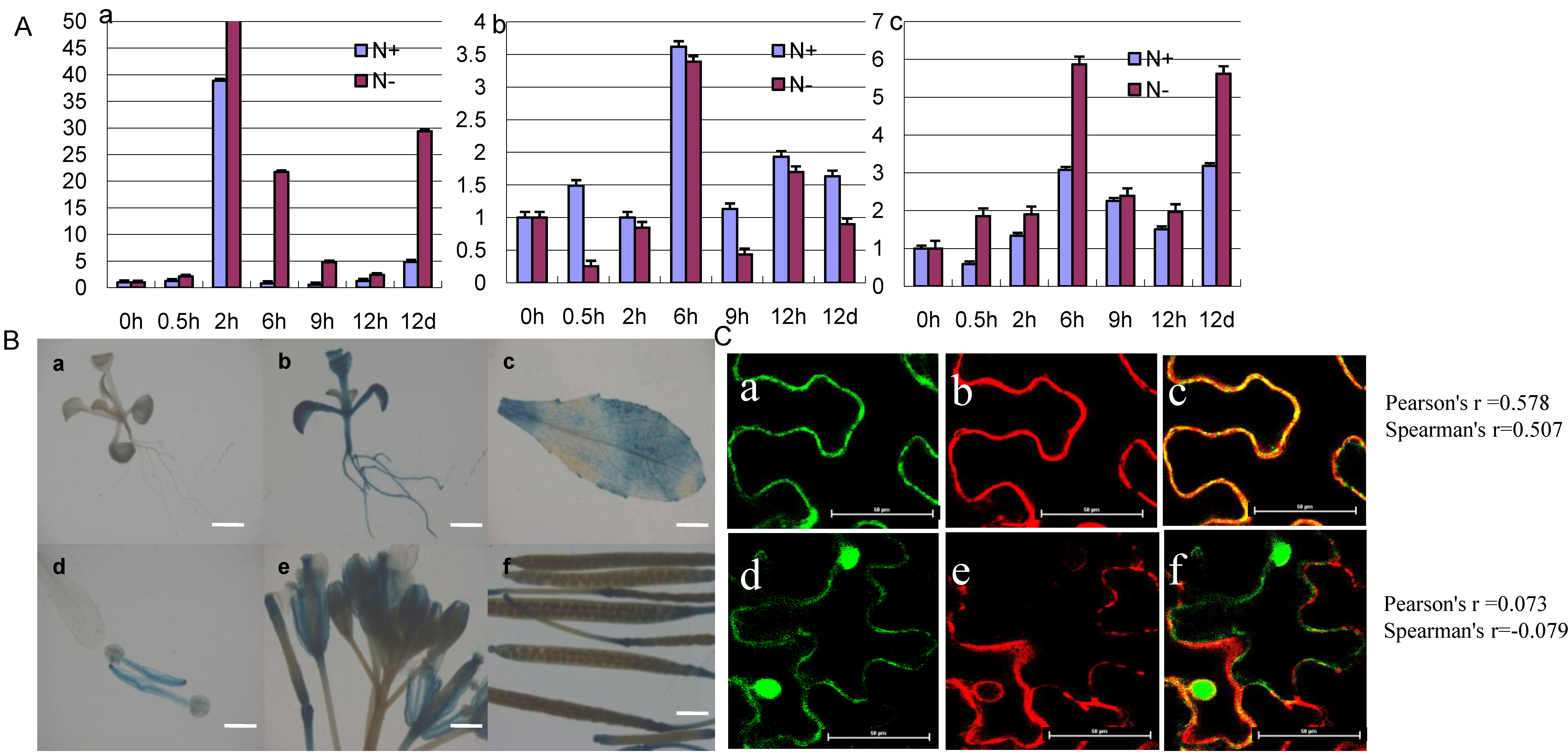
2.3. The Expression Pattern and Subcellular Localization of GmCBS21
2.4. Prokaryotic Expression and Cystathionine β-Synthase Activity Assay
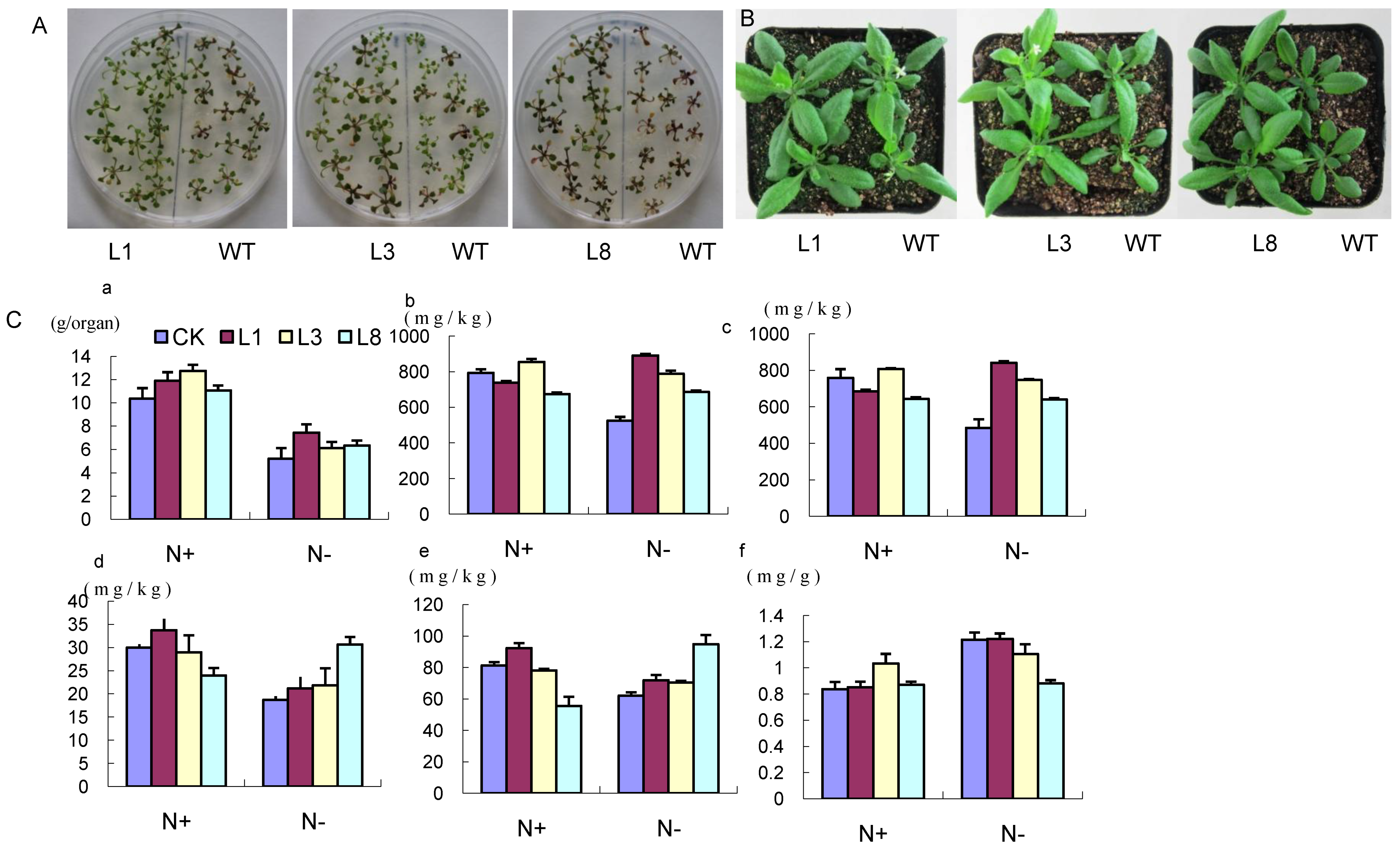
2.5. Phenotype of GmCBS21 Overexpression Transgenic Seedlings under Low Nitrogen Stress
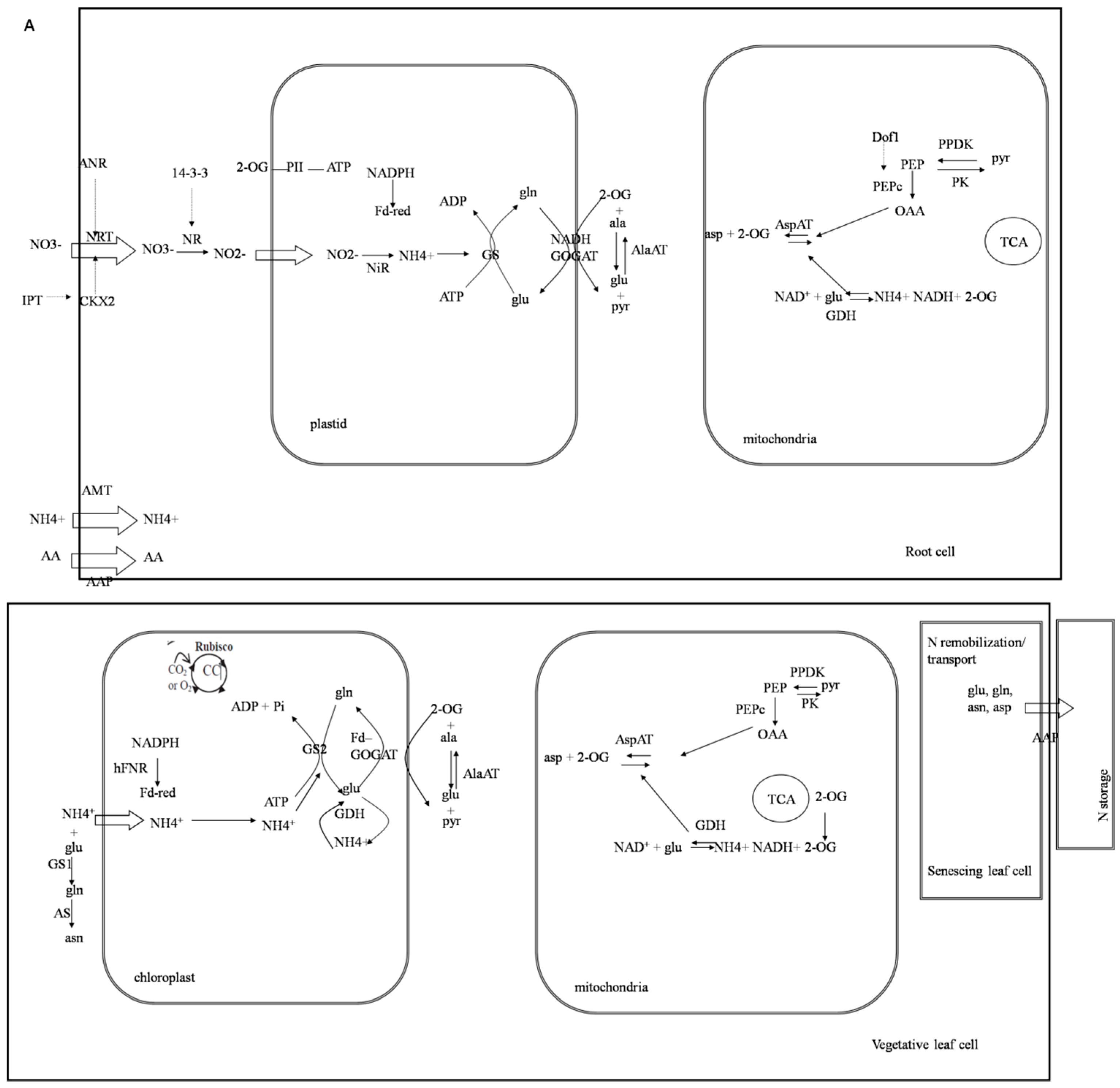
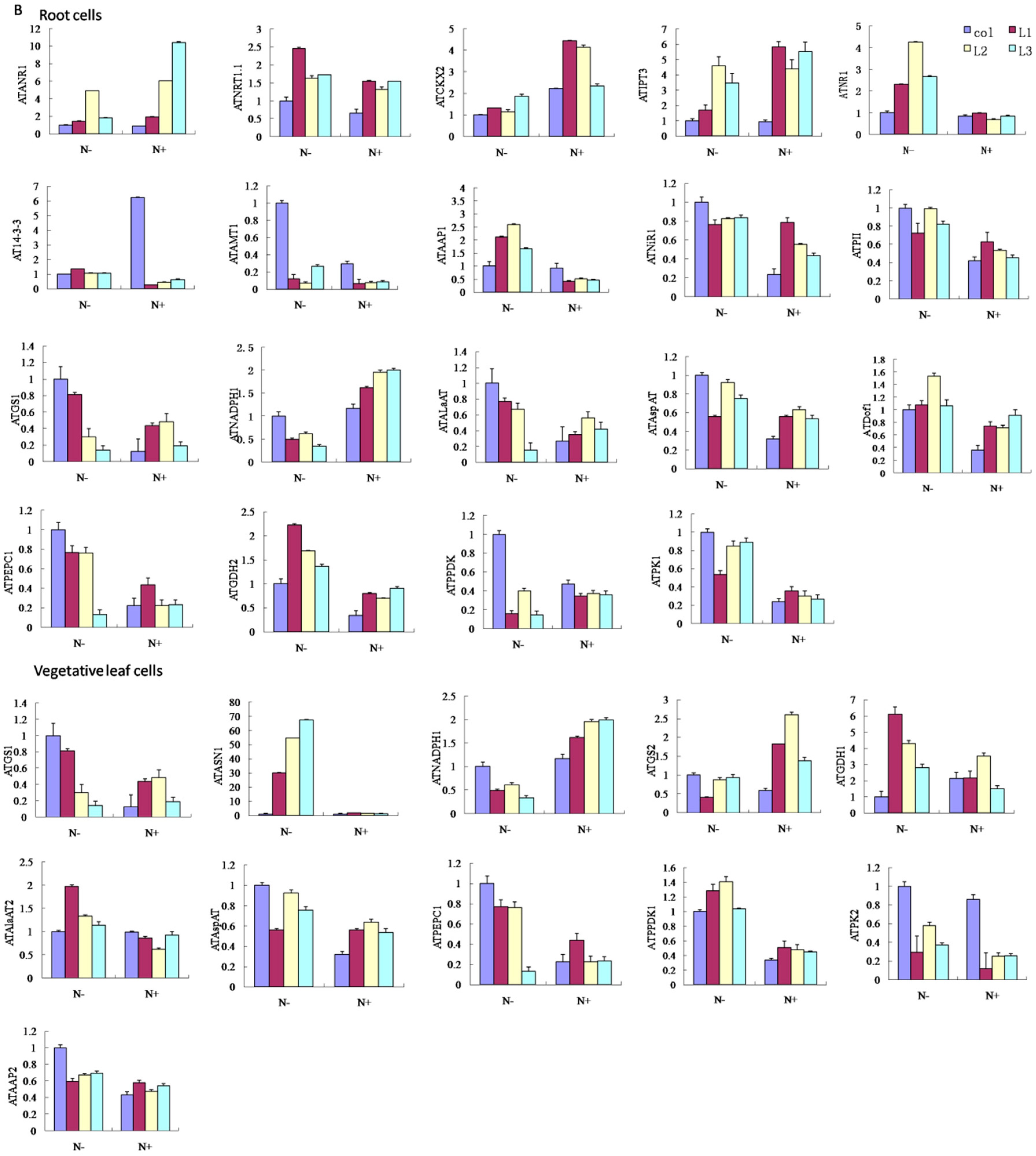
2.6. Physiological Indexes of GmCBS21 Overexpression Transgenic Seedlings under Low Nitrogen Stress
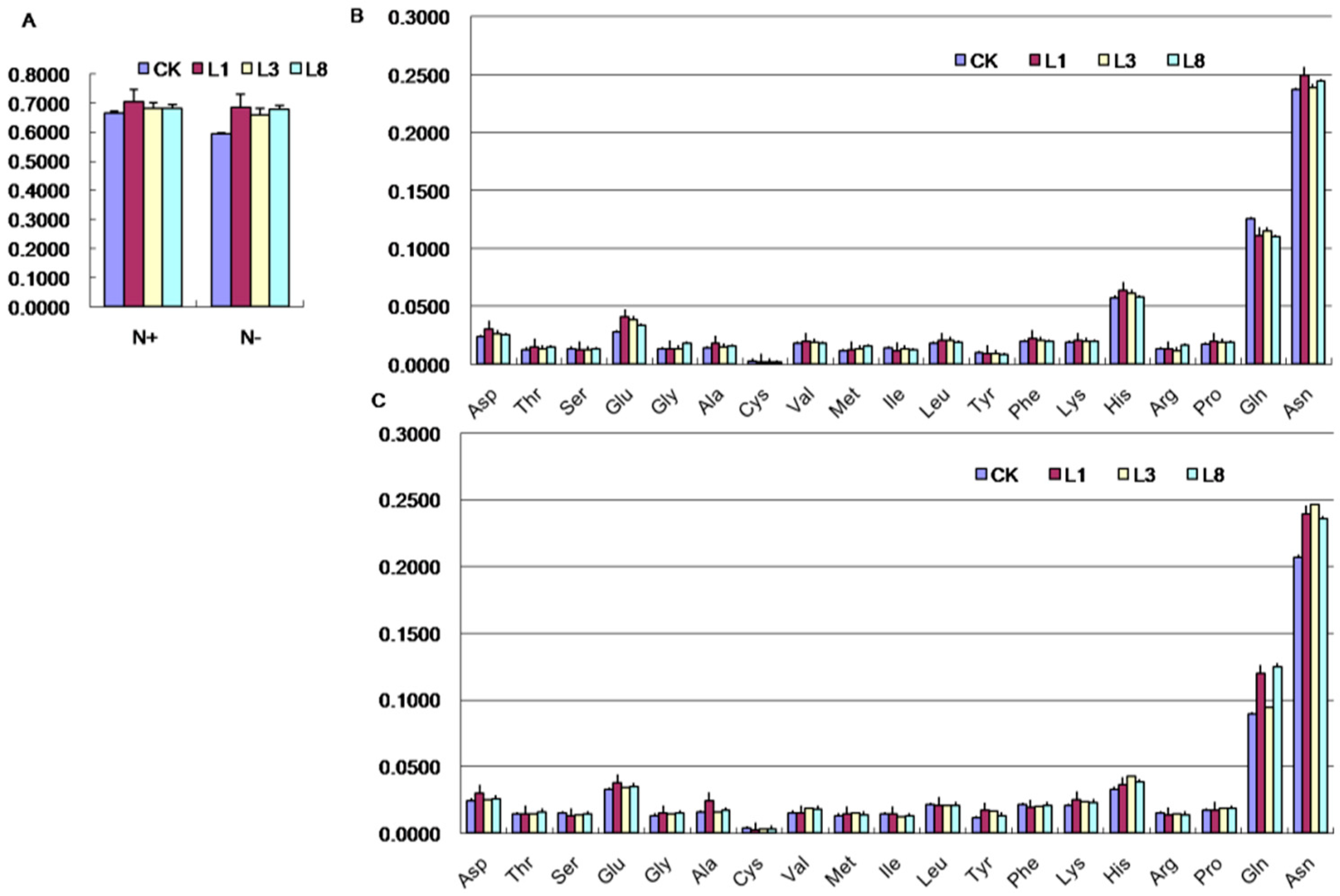
2.7. Effects of GmCBS21 Expression on Amino Acid Composition
3. Discussion
4. Materials and Methods
4.1. Database Searches for the Identification of CBS Domain-Containing Proteins Members in Soybean
4.2. Phylogenetic, Motif Recognition, Gene Structure and Chromosomal Location Analyses
4.3. Plant Materials and Growth Condition
4.4. RNA Extraction and Gene Expression Assays
4.5. Prokaryotic Expression and Cystathionine β-Synthase Activity Assay
4.6. Generation of GmCBS21 Transgenic Plants
4.7. Quantification of Nitrogen, Soluble Sugar, Amino Acids
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bao, A.; Liang, Z.; Zhao, Z.; Cai, H. Overexpressing of OsAMT1-3, a high affinity ammonium transporter gene, modifies rice growth and carbon-nitrogen metabolic status. Int. J. Mol. Sci. 2015, 16, 9037–9063. [Google Scholar] [CrossRef] [PubMed]
- Good, A.G.; Shrawat, A.K.; Muench, D.G. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 2004, 9, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Frink, C.R.; Waggoner, P.E.; Ausubel, J.H. Nitrogen fertilizer: Retrospect and prospect. Proc. Natl. Acad. Sci. USA 1999, 96, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.N.; Zhou, X.A.; Sha, A.H.; Wang, C.; Zhou, R.; Chen, S.L. Identification of genes associated with nitrogen-use efficiency by genome-wide transcriptional analysis of two soybean genotypes. BMC Genom. 2011, 12, 525. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997, 22, 12–13. [Google Scholar] [CrossRef]
- Sintchak, M.D.; Fleming, M.A.; Futer, O.; Raybuck, S.A.; Chambers, S.P.; Caron, P.R.; Murcko, M.A.; Wilson, K.P. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell 1996, 85, 921–930. [Google Scholar] [CrossRef]
- Schmidt-Rose, T.; Jentsch, T.J. Reconstitution of functional voltage-gated chloride channels from complementary fragments of CLC-1. J. Biol. Chem. 1997, 272, 20515–20521. [Google Scholar] [CrossRef] [PubMed]
- Woods, A.; Cheung, P.C.; Smith, F.C.; Davison, M.D.; Scott, J.; Beri, R.K.; Carling, D. Characterization of AMP-activated protein kinase beta and gamma subunits. Assembly of the heterotrimeric complex in vitro. J. Biol. Chem. 1996, 271, 10282–10290. [Google Scholar] [PubMed]
- Kemp, B.E. Bateman domains and adenosine derivatives form a binding contract. J. Clin. Investig. 2004, 113, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, H.R.; Singh, A.K.; Sopory, S.K.; Singla-Pareek, S.L.; Pareek, A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genom. 2009, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Ignoul, S.; Eggermont, J. CBS domains: Structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 2005, 289, C1369–C1378. [Google Scholar] [CrossRef] [PubMed]
- Ok, S.H.; Yoo, K.S.; Shin, J.S. CBSXs are sensor relay proteins sensing adenosine-containing ligands in Arabidopsis. Plant Signal. Behav. 2012, 7, 664–667. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.L.; Li, M.Y.; Liu, G.D.; Li, Y.D.; Sui, S.Z.; Guo, T.Y. Molecular characterization and functional prediction of a novel leaf SAG encoding a CBS-domain-containing protein from Coleus blumei. Chin. J. Biochem. Mol. Biol. 2007, 23, 249–255. [Google Scholar]
- Singh, A.K.; Kumar, R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Overexpression of rice CBS domain containing protein improves salinity, oxidative, and heavy metal tolerance in transgenic tobacco. Mol. Biotechnol. 2012, 52, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ren, X.; Zhu, L.; He, G. OsBi1, a rice gene, encodes a novel protein with a CBS-like domain and its expression is induced in responses to herbivore feeding. Plant Sci. 2004, 166, 1581–1588. [Google Scholar] [CrossRef]
- French, A.P.; Mills, S.; Swarup, R.; Bennett, M.J.; Pridmore, T.P. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 2008, 3, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Collart, F.R.; Osipiuk, J.; Trent, J.; Olsen, G.J.; Huberman, E. Cloning and characterization of the gene encoding IMP dehydrogenase from Arabidopsis thaliana. Gene 1996, 174, 217–220. [Google Scholar] [CrossRef]
- Emanuelle, S.; Hossain, M.I.; Moller, I.E.; Pedersen, H.L.; van de Meene, A.M.; Doblin, M.S.; Koay, A.; Oakhill, J.S.; Scott, J.W.; Willats, W.G.; et al. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015, 82, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.D.; Tang, R.J.; Liu, H.; Gao, X.S.; Zheng, H.Q.; Zhang, H.X. Cloning and molecular analyses of the Arabidopsis thaliana chloride channel gene family. Plant Sci. 2009, 176, 650–661. [Google Scholar] [CrossRef]
- Hechenberger, M.; Schwappach, B.; Fischer, W.N.; Frommer, W.B.; Jentsch, T.J.; Steinmeyer, K. A family of putative chloride channels from Arabidopsis and functional complementation of a yeast strain with a CLC gene disruption. J. Biol. Chem. 1996, 271, 33632–33638. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Su, S.Z.; Wu, Y.; Li, S.P.; Shan, X.H.; Liu, H.K.; Wang, S.; Yuan, Y.P. Overexpression of maize chloride channel gene ZmCLC-D in Arabidopsis thaliana improved its stress resistance. Biol. Plant. 2015, 59, 55–64. [Google Scholar] [CrossRef]
- Cao, Y.; Schubert, K.R. Molecular cloning and characterization of a cDNA encoding soybean nodule IMP dehydrogenase. Biochim. Biophys. Acta 2001, 1520, 242–246. [Google Scholar] [CrossRef]
- Li, W.Y.; Wong, F.L.; Tsai, S.N.; Phang, T.H.; Shao, G.; Lam, H.M. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ. 2006, 29, 1122–1137. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.H.; Li, M.W.; Yao, X.Q.; Lam, H.M. The GmCLC1 protein from soybean functions as a chloride ion transporter. J. Plant Physiol. 2013, 170, 101–104. [Google Scholar] [CrossRef] [PubMed]
- McAllister, C.H.; Beatty, P.H.; Good, A.G. Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 2012, 10, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Kudo, T.; Kojima, M.; Sakakibara, H. Hormonal control of nitrogen acquisition: Roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 2011, 62, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Ueda, N.; Aoki, K.; Kuromori, T.; Hirayama, T.; Shinozaki, K.; Yamaya, T.; Sakakibara, H. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 2004, 45, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.M.; Wong, P.; Chan, H.K.; Yam, K.M.; Chen, L.; Chow, C.M.; Coruzzi, G.M. Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 2003, 132, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Coruzzi, G.M.; Zhou, L. Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects”. Curr. Opin. Plant Biol. 2001, 4, 247–253. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 2000, 21, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Ferrario-Mery, S.; Besin, E.; Pichon, O.; Meyer, C.; Hodges, M. The regulatory PII protein controls arginine biosynthesis in Arabidopsis. FEBS Lett. 2006, 580, 2015–2020. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.; Nunes-Nesi, A.; Parsley, K.; Leiss, A.; Leach, G.; Coates, S.; Wingler, A.; Fernie, A.R.; Hibberd, J.M. Cytosolic pyruvate, orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. Plant J. 2010, 62, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Rolletschek, H.; Hosein, F.; Miranda, M.; Heim, U.; Gotz, K.P.; Schlereth, A.; Borisjuk, L.; Saalbach, I.; Wobus, U.; Weber, H. Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 2005, 137, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, Y.; Dolferus, R.; Ismond, K.P.; Good, A.G. Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J. 2007, 49, 1108–1121. [Google Scholar] [CrossRef] [PubMed]
- Hawkesford, M.J.; Barraclough, P. Biotechnological Approaches to Improving Nitrogen Use Efficiency in Plants: Alanine Aminotransferase as a Case Study; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Shrawat, A.K.; Carroll, R.T.; DePauw, M.; Taylor, G.J.; Good, A.G. Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 2008, 6, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, Y.; Good, A.G. NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J. Exp. Bot. 2008, 59, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis genome database, AIR 9.0. Avaliable online: http://www.arabidopsis.org/ (accessed on 1 March 2016).
- Rice Genome Annotation Project. Avaliable online: http://rice.plantbiology.msu.edu/index.shtml (accessed on 1 March 2016).
- Phytozome (v9.0) database. Avaliable online: http://www.phytozome.net (accessed on 1 March 2016).
- The Pfam database. Avaliable online: http://Pfam.sanger.ac.uk/ (accessed on 1 March 2016).
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- The Gene Indices. Avaliable online: http://compbio.dfci.harvard.edu/tgi/ (accessed on 1 March 2016).
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan 2007, 29, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Cannon, E.K.; Cannon, S.B. Chromosome visualization tool: A whole genome viewer. Int. J. Plant Genom. 2011, 2011, 373875. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Nakagawa, H. Studies on cystathionine synthetase characteristics of purified rat liver enzyme. J. Biochem. 1971, 69, 711–723. [Google Scholar] [PubMed]
- Davis, A.M.; Hall, A.; Millar, A.J.; Darrah, C.; Davis, S.J. Protocol: Streamlined sub-protocols for floral-DIP transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 2009, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Kimberly, A.E.; Roberts, M.G. A method for the direct determination of organic nitrogen by the kjeldahl process. Public Health Pap. Rep. 1905, 31, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Geiger, M.; Walch-Liu, P.; Engels, C.; Harnecker, J.; Schulze, E.-D.; Ludewig, F.; Sonnewald, U.; Scheible, W.-R.; Stitt, M. Enhanced carbon dioxide leads to a modified diurnal rhythm of nitrate reductase activity in older plants, and a large stimulation of nitrate reductase activity. Plant Cell Environ. 1998. [Google Scholar] [CrossRef]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Q.; Shang, W.; Zhang, C.; Chen, H.; Chen, L.; Yuan, S.; Chen, S.; Zhang, X.; Zhou, X. Identification and Comparative Analysis of CBS Domain-Containing Proteins in Soybean (Glycine max) and the Primary Function of GmCBS21 in Enhanced Tolerance to Low Nitrogen Stress. Int. J. Mol. Sci. 2016, 17, 620. https://doi.org/10.3390/ijms17050620
Hao Q, Shang W, Zhang C, Chen H, Chen L, Yuan S, Chen S, Zhang X, Zhou X. Identification and Comparative Analysis of CBS Domain-Containing Proteins in Soybean (Glycine max) and the Primary Function of GmCBS21 in Enhanced Tolerance to Low Nitrogen Stress. International Journal of Molecular Sciences. 2016; 17(5):620. https://doi.org/10.3390/ijms17050620
Chicago/Turabian StyleHao, Qingnan, Weijuan Shang, Chanjuan Zhang, Haifeng Chen, Limiao Chen, Songli Yuan, Shuilian Chen, Xiaojuan Zhang, and Xinan Zhou. 2016. "Identification and Comparative Analysis of CBS Domain-Containing Proteins in Soybean (Glycine max) and the Primary Function of GmCBS21 in Enhanced Tolerance to Low Nitrogen Stress" International Journal of Molecular Sciences 17, no. 5: 620. https://doi.org/10.3390/ijms17050620






