Gastrointestinal Endogenous Protein-Derived Bioactive Peptides: An in Vitro Study of Their Gut Modulatory Potential
Abstract
:1. Introduction
2. Results
2.1. Digestion of Proteins and Determination of Protein Content Using Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis SDS-PAGE
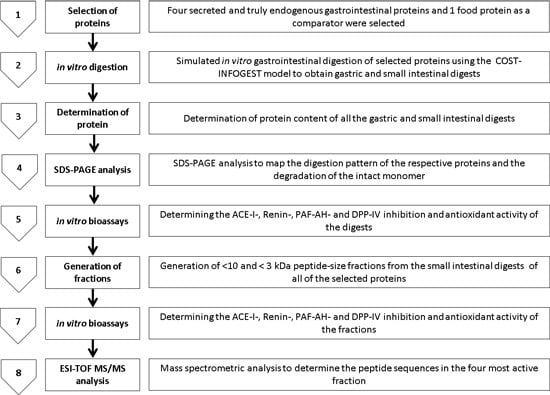
2.2. Angiotensin-I Converting Enzyme (ACE-I) Inhibition by Digests and Enriched Fractions
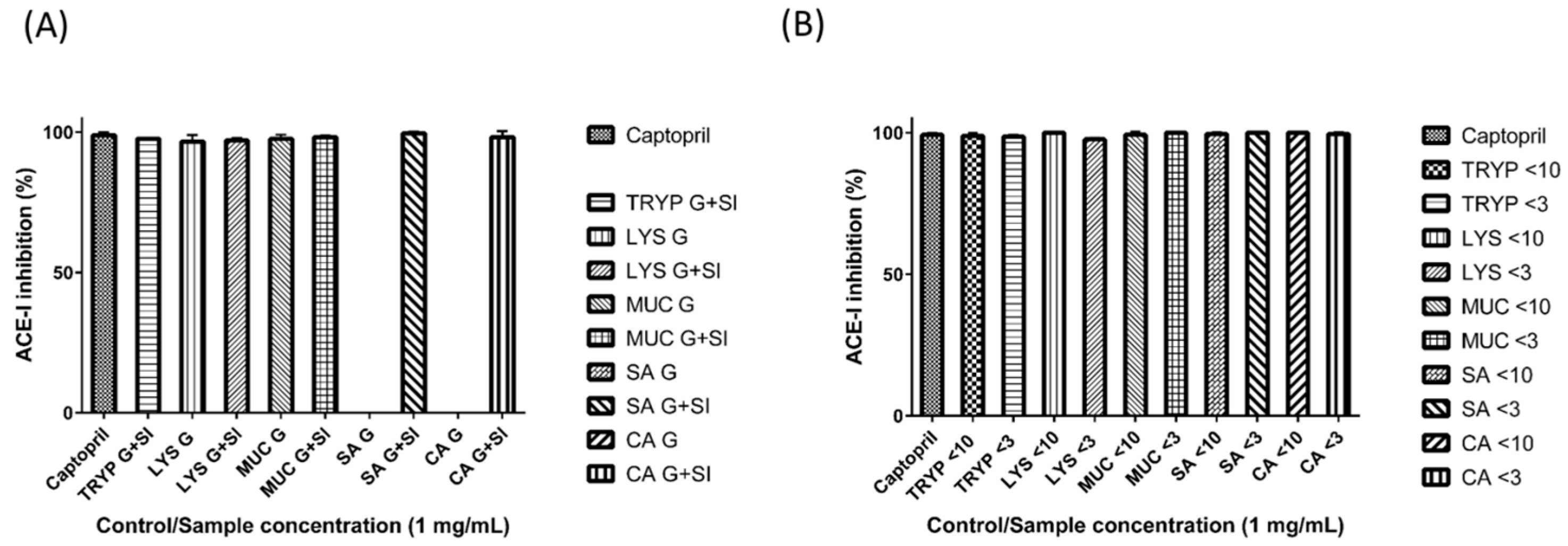
2.3. Renin Inhibition by Digests and Enriched Fractions
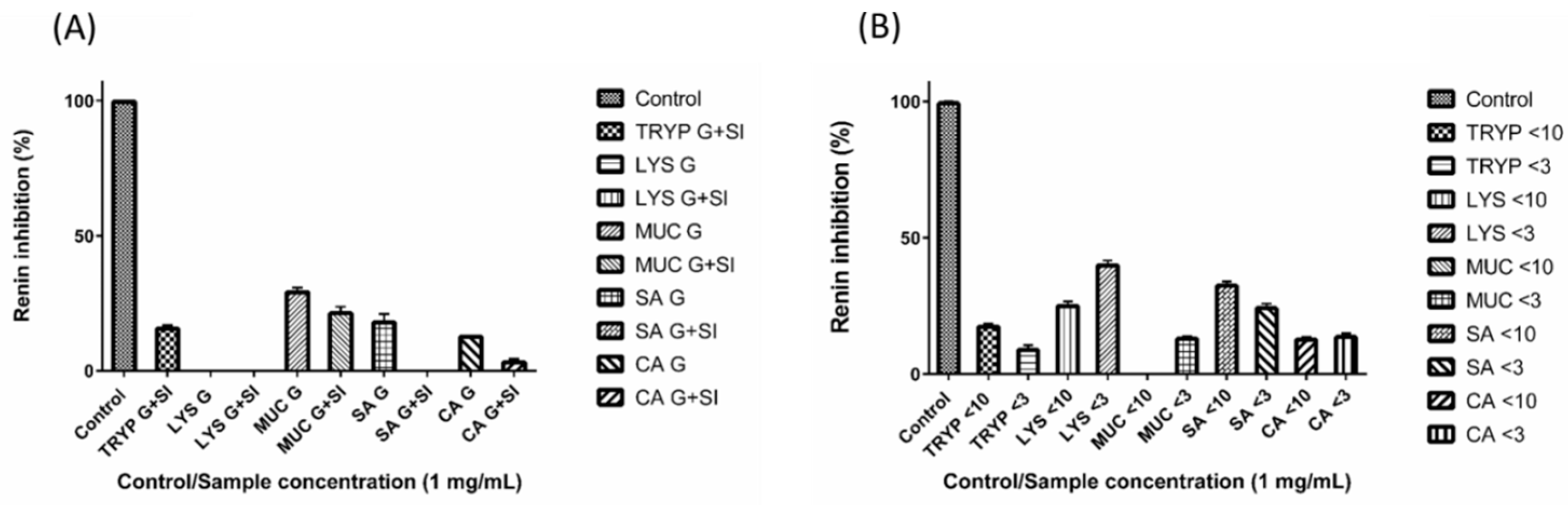
2.4. Renin, Platelet-Activating Factor-Acetylhydrolase (PAF-AH) Inhibition by Digests and Enriched Fractions
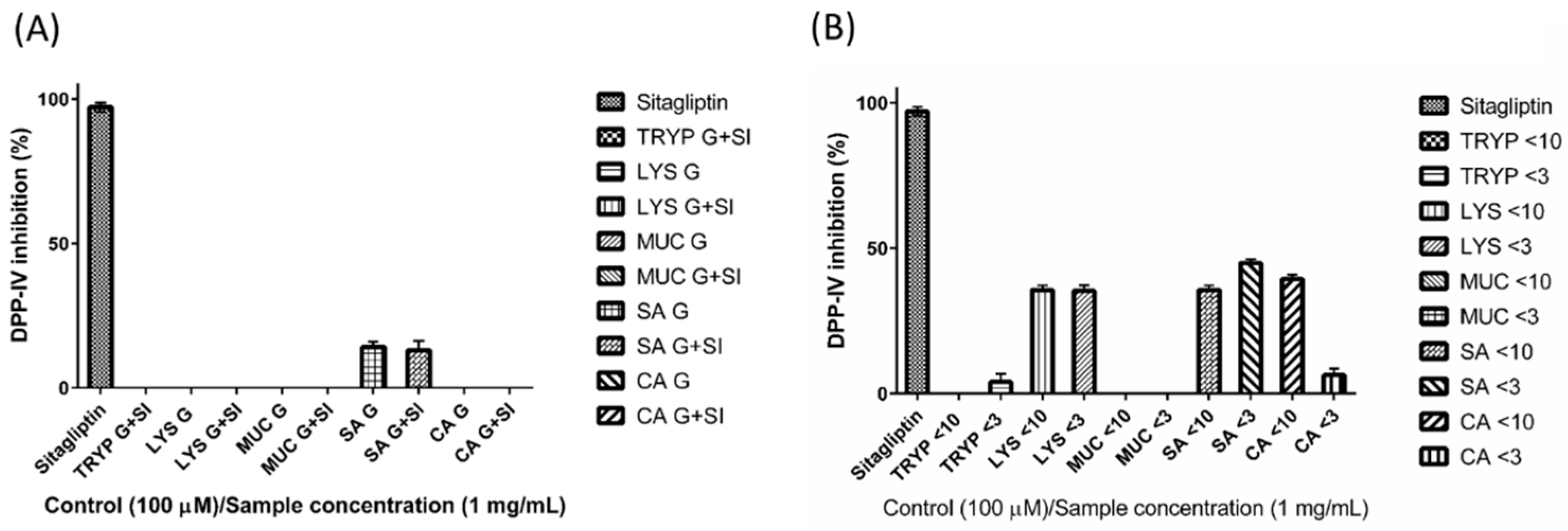
2.5. Dipeptidyl Peptidase-IV Inhibitory (DPP-IV) Inhibition by Digests and Enriched Fractions
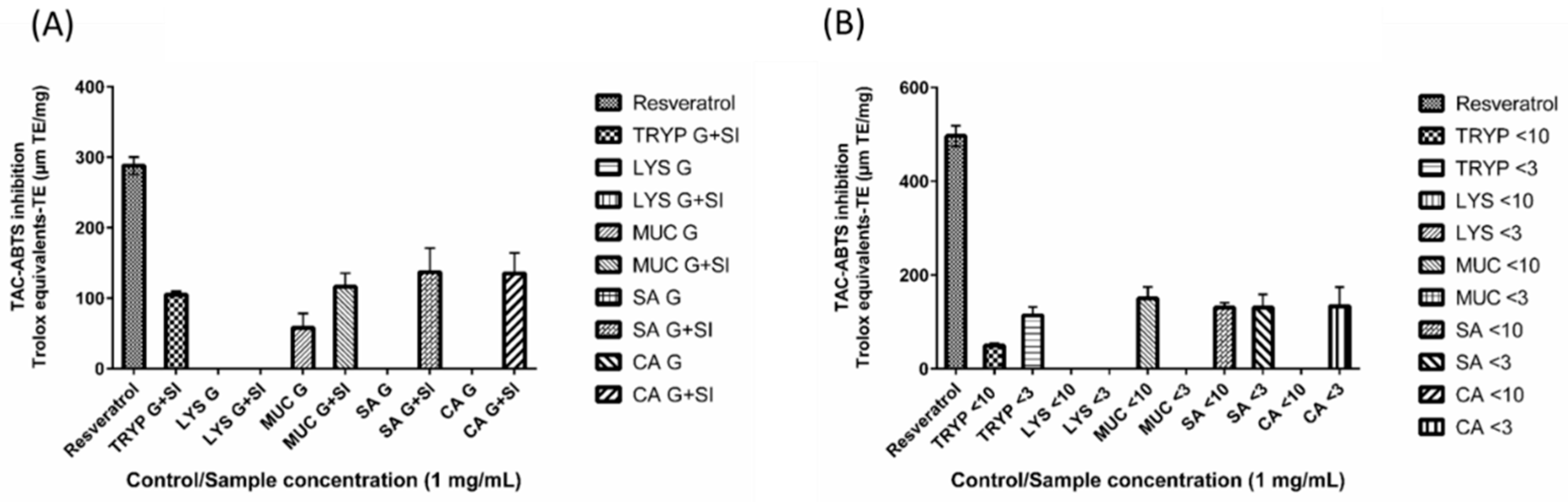
2.6. 2,2′-Azino-bis-3-ethylbenzthiazoline-6-sulphonic Acid Total Antioxidant Capacity (ABTS-TAC) of Digests and Enriched Fractions
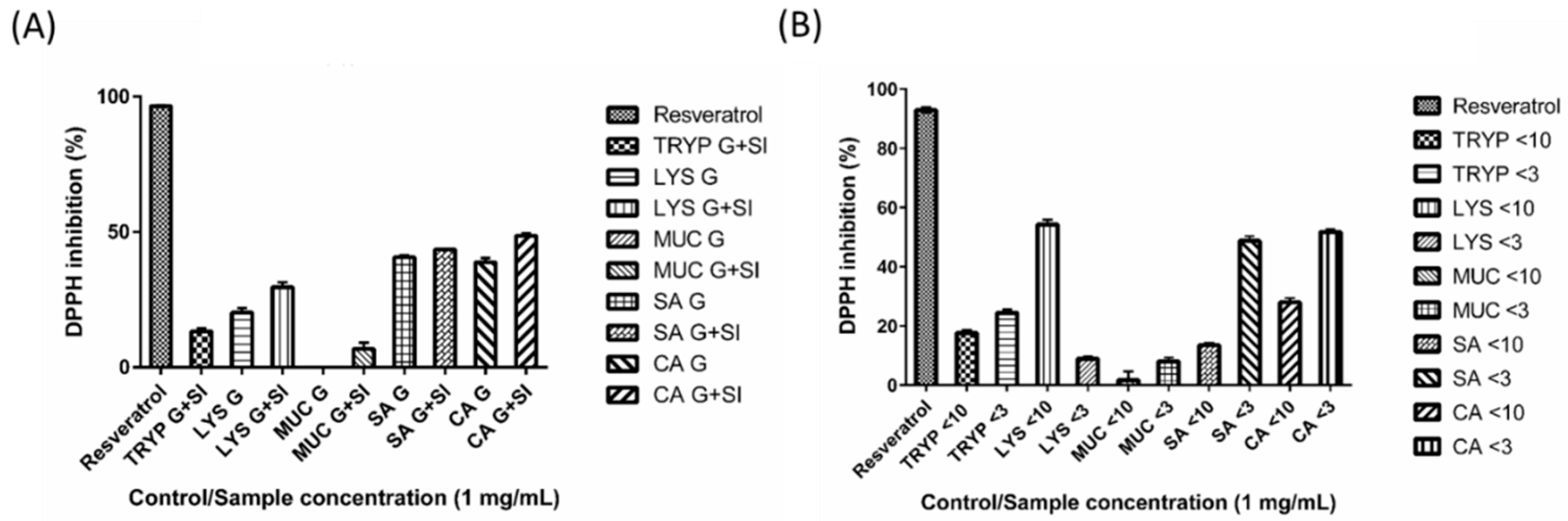
2.7. DPPH Inhibition by Digests and Enriched Fractions
2.8. Electrospray Ionisation Time of Flight Mass Spectrometry (ESI-TOF-MS) Characterisation of Peptides
3. Discussion
4. Experimental Section
4.1. Materials and Reagents
4.2. Simulated Gastrointestinal Digestion of Selected Gastrointestinal Endogenous Proteins (GEP) and a Food Protein Comparator
4.3. Total Protein Analysis
4.4. Tris Tricine Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS-PAGE) Analysis
4.5. ACE-I Inhibition Assay
4.6. Renin Inhibition Assay
4.7. PAF-AH Inhibition Assay
4.8. DPP-IV Inhibition
4.9. 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS)-Based Total Antioxidant Capacity (ABTS-TAC) Assay
4.10. DPPH Inhibition
4.11. Electrospray Ionization Time of Flight Mass Spectrometry (ESI-TOF MS) Characterisation of Peptides Present in the Most Active Fractions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABTS | 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) |
| ACE-I | angiotensin-I Converting Enzyme |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| DPP-IV | dipeptidyl peptidase-IV |
| GEP | Gastrointestinal endogenous proteins |
| GLP-2 | Glucagon-like peptide-2 |
| MAFP | Methyl arachidonyl fluorophosphonate |
| MWCO | Molecular weight cut-off |
| PAF-AH | platelet-activating factor-acetylhydrolase |
| RAAS | renin angiotensin aldosterone system |
| SDS-PAGE | sodium dodecyl sulphate polyacrylamide gel electrophoresis |
References
- Moughan, P.J.; Rutherfurd, S.M.; Montoya, C.A.; Dave, L.A. Food-derived bioactive peptides—A new paradigm. Nutr. Res. Rev. 2014, 27, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Dave, L.A.; Hayes, M.; Montoya, C.A.; Rutherfurd, S.M.; Moughan, P.J. Human gut endogenous proteins as a potential source of angiotensin-I-converting enzyme (ACE-I)-, renin inhibitory and antioxidant peptides. Peptides 2016, 76, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Dave, L.A.; Montoya, C.A.; Rutherfurd, S.M.; Moughan, P.J. Gastrointestinal endogenous proteins as a source of bioactive peptides—An in silico study. PLoS ONE 2014, 9, e98922. [Google Scholar] [CrossRef] [PubMed]
- Miner-Williams, W.; Moughan, P.J.; Fuller, M.F. Endogenous components of digesta protein from the terminal ileum of pigs fed a casein-based diet. J. Agric. Food Chem. 2009, 57, 2072–2078. [Google Scholar] [CrossRef] [PubMed]
- Moughan, P.J.; Rutherfurd, S.M. Gut luminal endogenous protein: Implications for the determination of ileal amino acid digestibility in humans. Br. J. Nutr. 2012, 108 (Suppl. S2), S258–S263. [Google Scholar] [CrossRef] [PubMed]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carriere, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, D.A. What if gut hormones aren’t really hormones: DPP-4 inhibition and local action of GLP-1 in the gastrointestinal tract. Endocrinology 2011, 152, 2925–2926. [Google Scholar] [CrossRef] [PubMed]
- Pucar, L.B.; Detel, D.; Varljen, J. Dipeptidyl peptidase IV in inflammatory bowel diseases (DPP IV/CD26). Arhiv. Ind. Hyg. Toxicol. 2012, 63, 75–100. [Google Scholar]
- Carl-McGrath, S.; Grantzdorffer, I.; Lendeckel, U.; Ebert, M.P.; Rocken, C. Angiotensin II-generating enzymes, angiotensin-converting enzyme (ACE) and mast cell chymase (CMA1), in gastric inflammation may be regulated by H. Pylori and associated cytokines. Pathology 2009, 41, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Kanasaki, K.; Shi, S.; Kanasaki, M.; He, J.; Nagai, T.; Nakamura, Y.; Ishigaki, Y.; Kitada, M.; Srivastava, S.P.; Koya, D. Linagliptin-mediated DPP-4 inhibition ameliorates kidney fibrosis in streptozotocin-induced diabetic mice by inhibiting endothelial-to-mesenchymal transition in a therapeutic regimen. Diabetes 2014, 63, 2120–2131. [Google Scholar] [CrossRef] [PubMed]
- Coates, D. The angiotensin converting enzyme (ACE). Int. J. Biochem. Cell Biol. 2003, 35, 769–773. [Google Scholar] [CrossRef]
- Fandriks, L. The renin-angiotensin system and the gastrointestinal mucosa. Acta Physiol. (Oxf.) 2011, 201, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Garg, M.; Angus, P.W.; Burrell, L.M.; Herath, C.; Gibson, P.R.; Lubel, J.S. Review article: The pathophysiological roles of the renin-angiotensin system in the gastrointestinal tract. Aliment. Pharmacol. Ther. 2012, 35, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tan, X.-D.; Qu, X.-W.; Chang, H.; Remick, D.G.; Gonzalez-Crussi, F.; Hsueh, W. Platelet-activating factor (PAF) up-regulates plasma and tissue PAF-acetylhydrolase activity in the rat: Effect of cycloheximide. Pediatr. Res. 1997, 42, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Packard, C.J.; O’Reilly, D.S.J.; Caslake, M.J.; McMahon, A.D.; Ford, I.; Cooney, J.; Macphee, C.H.; Suckling, K.E.; Krishna, M.; Wilkinson, F.E.; et al. Lipoprotein-associated phospholipase a2 as an independent predictor of coronary heart disease. N. Engl. J. Med. 2000, 343, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Sudhir, K. Lipoprotein-associated phospholipase A2, a novel inflammatory biomarker and independent risk predictor for cardiovascular disease. J. Clin. Endocrinol. Metab. 2005, 90, 3100–3105. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, P.; Zhang, L.; Osman, H.; Mohler Iii, E.R.; Macphee, C.; Zalewski, A.; Postle, A.; Wilensky, R.L. Role of lipoprotein-associated phospholipase A2 in leukocyte activation and inflammatory responses. Atherosclerosis 2007, 191, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Okawada, M.; Holst, J.J.; Teitelbaum, D.H. Administration of a DPP-IV inhibitor enhances the intestinal adaptation in a mouse model of short bowel syndrome. Surgery 2011, 150, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Salles, T.A.; dos Santos, L.; Barauna, V.G.; Girardi, A.C. Potential role of dipeptidyl peptidase IV in the pathophysiology of heart failure. Int. J. Mol. Sci. 2015, 16, 4226–4249. [Google Scholar] [CrossRef] [PubMed]
- Reinehr, T.; Roth, C.L.; Enriori, P.J.; Masur, K. Changes of dipeptidyl peptidase IV (DPP-IV) in obese children with weight loss: Relationships to peptide YY, pancreatic peptide, and insulin sensitivity. J. Pediatr. Endocrinol. Metab. 2010, 23, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Zhao, K.; Whiteman, M. The gastrointestinal tract: A major site of antioxidant action? Free Radic. Res. 2000, 33, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, S.M.; Moughan, P.J.; Reynolds, G.W.; James, K.A. The effect of dietary peptide concentration on endogenous ileal amino acid loss in the growing pig. Br. J. Nutr. 2000, 83, 421–430. [Google Scholar] [PubMed]
- Montagne, L.; Toullec, R.; Lalles, J.P. Intestinal digestion of dietary and endogenous proteins along the small intestine of calves fed soybean or potato. J. Anim. Sci. 2001, 79, 2719–2730. [Google Scholar] [PubMed]
- Baglieri, A.; Mahe, S.; Benamouzig, R.; Savoie, L.; Tome, D. Digestion patterns of endogenous and different exogenous proteins affect the composition of intestinal effluents in humans. J. Nutr. 1995, 125, 1894–1903. [Google Scholar] [PubMed]
- Miner-Williams, W.; Deglaire, A.; Benamouzig, R.; Fuller, M.F.; Tome, D.; Moughan, P.J. Endogenous proteins in terminal ileal digesta of adult subjects fed a casein-based diet. Am. J. Clin. Nutr. 2012, 96, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Miner-Williams, W.; Deglaire, A.; Benamouzig, R.; Fuller, M.F.; Tomé, D.; Moughan, P.J. Endogenous proteins in the ileal digesta of adult humans given casein-, enzyme-hydrolyzed casein-or crystalline amino-acid-based diets in an acute feeding study. Eur. J. Nutr. 2014, 68, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Deglaire, A.; Moughan, P.J.; Rutherfurd, S.M.; Bos, C.; Tome, D. Feeding dietary peptides to growing rats enhances gut endogenous protein flows compared with feeding protein-free or free amino acid-based diets. J. Nutr. 2007, 137, 2431–2436. [Google Scholar] [PubMed]
- Gilani, G.S.; Cockell, K.A.; Sepehr, E. Effects of antinutritional factors on protein digestibility and amino acid availability in foods. J. AOAC Int. 2005, 88, 967–987. [Google Scholar] [PubMed]
- Rutherfurd, S.M.; Cui, J.; Goroncy, A.K.; Moughan, P.J. Dietary protein structure affects endogenous ileal amino acids but not true ileal amino acid digestibility in growing male rats. J. Nutr. 2015, 145, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, D.R.; Demers, M.; Zuur, G.; Lobley, G.E.; Seoane, J.R.; Nolan, J.V.; Lapierre, H. Effect of dietary fiber on endogenous nitrogen flows in lactating dairy cows. J. Dairy Sci. 2002, 85, 3013–3025. [Google Scholar] [CrossRef]
- Rutherfurd, S.M.; Fanning, A.C.; Miller, B.J.; Moughan, P.J. Protein digestibility-corrected amino acid scores and digestible indispensable amino acid scores differentially describe protein quality in growing male rats. J. Nutr. 2015, 145, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jiménez-Saiz, R.; Benedé, S.; Miralles, B.; López-Expósito, I.; Molina, E.; López-Fandiño, R. Immunological behavior of in vitro digested egg-white lysozyme. Mol. Nutr. Food Res. 2014, 58, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-originating ace inhibitors, including antihypertensive peptides, as preventive food components in blood pressure reduction. Compr. Rev. Food Sci. Food Saf. 2014, 13, 114–134. [Google Scholar] [CrossRef]
- BIOPEP. Available online: http://www.uwm.edu.pl/biochemia/index.php/en/biopep (accessed on 25 August 2015).
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. Biopep database and other programs for processing bioactive peptide sequences. J. AOAC Int. 2008, 91, 965–980. [Google Scholar] [PubMed]
- Kvietys, P.R. Chapter 5: Postprandial hyperemia. In The Gastrointestinal Circulation; Morgan & Claypool Life Sciences: San Rafael, CA, USA, 2010. [Google Scholar]
- Kvietys, P.R.; McLendon, J.M.; Granger, D.N. Postprandial intestinal hyperemia: Role of bile salts in the ileum. Am. J. Physiol. Gastrointest. Liver Physiol. 1981, 241, G469–G477. [Google Scholar]
- Fitzgerald, C.; Mora-Soler, L.; Gallagher, E.; O’Connor, P.; Prieto, J.; Soler-Vila, A.; Hayes, M. Isolation and characterization of bioactive pro-peptides with in vitro renin inhibitory activities from the macroalga palmaria palmata. J. Agric. Food Chem. 2012, 60, 7421–7427. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C.; Gallagher, E.; O’Connor, P.; Prieto, J.; Mora-Soler, L.; Grealy, M.; Hayes, M. Development of a seaweed derived platelet activating factor acetylhydrolase (PAF-AH) inhibitory hydrolysate, synthesis of inhibitory peptides and assessment of their toxicity using the zebrafish larvae assay. Peptides 2013, 50, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yu, G.; Zhang, Y.; Zhang, B.; Fan, J. Dipeptidyl peptidase IV inhibitory peptides derived from oat (avena sativa L.), buckwheat (fagopyrum esculentum), and highland barley (hordeum vulgare trifurcatum (L.) trofim) proteins. J. Agric. Food Chem. 2015, 63, 9543–9549. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Higashiyama, M.; Kaji, I.; Rudenkyy, S.; Higuchi, K.; Guth, P.H.; Engel, E.; Kaunitz, J.D.; Akiba, Y. Dipeptidyl peptidase IV inhibition prevents the formation and promotes the healing of indomethacin-induced intestinal ulcers in rats. Dig. Dis. Sci. 2014, 59, 1286–1295. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Wang, J.-H.; Higashiyama, M.; Rudenkyy, S.; Higuchi, K.; Guth, P.H.; Engel, E.; Kaunitz, J.D.; Akiba, Y. Dipeptidyl peptidase IV inhibition potentiates amino acid- and bile acid-induced bicarbonate secretion in rat duodenum. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G810–G816. [Google Scholar] [CrossRef] [PubMed]
- Sueyoshi, R.; Woods Ignatoski, K.M.; Okawada, M.; Hartmann, B.; Holst, J.; Teitelbaum, D.H. Stimulation of intestinal growth and function with DPP4 inhibition in a mouse short bowel syndrome model. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G410–G419. [Google Scholar] [CrossRef] [PubMed]
- Lafarga, T.; Rai, D.K.; O’Connor, P.; Hayes, M. A bovine fibrinogen-enriched fraction as a source of peptides with in vitro renin and angiotensin-i-converting enzyme inhibitory activities. J. Agric. Food Chem. 2015, 63, 8676–8684. [Google Scholar] [CrossRef] [PubMed]
- Darewicz, M.; Borawska, J.; Vegarud, G.E.; Minkiewicz, P.; Iwaniak, A. Angiotensin I-converting enzyme (ACE) inhibitory activity and ACE inhibitory peptides of salmon (salmo salar) protein hydrolysates obtained by human and porcine gastrointestinal enzymes. Int. J. Mol. Sci. 2014, 15, 14077–14101. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yan, J.; Yu, Y.; Qi, Y. Screening and identification of DPP-IV inhibitory peptides from deer skin hydrolysates by an integrated approach of lC–MS/MS and in silico analysis. J. Funct. Foods Part A 2015, 18, 344–357. [Google Scholar] [CrossRef]
- Atlas, S.A. The renin-angiotensin aldosterone system: Pathophysiological role and pharmacologic inhibition. J. Manag. Care Pharm. 2007, 13, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Samsamshariat, S.; Basati, G.; Movahedian, A.; Pourfarzam, M.; Sarrafzadegan, N. Elevated plasma platelet-activating factor acetylhydrolase activity and its relationship to the presence of coronary artery disease. J. Res. Med. Sci. 2011, 16, 674–679. [Google Scholar] [PubMed]
- Hernández-Ledesma, B.; Quirós, A.; Amigo, L.; Recio, I. Identification of bioactive peptides after digestion of human milk and infant formula with pepsin and pancreatin. Int. Dairy J. 2007, 17, 42–49. [Google Scholar] [CrossRef]
- Kambayashi, Y.; Binh, N.T.; Asakura, H.W.; Hibino, Y.; Hitomi, Y.; Nakamura, H.; Ogino, K. Efficient assay for total antioxidant capacity in human plasma using a 96-well microplate. J. Clin. Biochem. Nutr. 2009, 44, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Moller, N.P.; Scholz-Ahrens, K.E.; Roos, N.; Schrezenmeir, J. Bioactive peptides and proteins from foods: Indication for health effects. Eur. J. Nutr. 2008, 47, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Zhong, Y. Bioactive peptides. J. AOAC Int. 2008, 91, 914–931. [Google Scholar] [PubMed]
- Rajapakse, N.; Mendis, E.; Byun, H.G.; Kim, S.K. Purification and in vitro antioxidative effects of giant squid muscle peptides on free radical-mediated oxidative systems. J. Nutr. Biochem. 2005, 16, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Girgih, A.T.; Alashi, A.M.; He, R.; Malomo, S.A.; Raj, P.; Netticadan, T.; Aluko, R.E. A novel hemp seed meal protein hydrolysate reduces oxidative stress factors in spontaneously hypertensive rats. Nutrition 2014, 6, 5652–5666. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.-F.; Hu, F.-Y.; Wang, B.; Li, Z.-R.; Luo, H.-Y. Influence of amino acid compositions and peptide profiles on antioxidant capacities of two protein hydrolysates from skipjack tuna (katsuwonus pelamis) dark muscle. Mar. Drugs 2015, 13, 2580–2601. [Google Scholar] [CrossRef] [PubMed]
- Kitts, D.D.; Weiler, K. Bioactive proteins and peptides from food sources. Applications of bioprocesses used in isolation and recovery. Curr. Pharm. Des. 2003, 9, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Muramoto, K.; Yamauchi, F. Structural analysis of antioxidative peptides from soybean β-conglycinin. J. Agric. Food Chem. 1995, 43, 574–578. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Chemometric analysis of the amino acid requirements of antioxidant food protein hydrolysates. Int. J. Mol. Sci. 2011, 12, 3148–3161. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: Quantitative structure-activity relationship study of di- and tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Sokołowska, J.; Starowicz, P.; Bucholska, J.; Hrynkiewicz, M. Common amino acid subsequences in a universal proteome—Relevance for food science. Int. J. Mol. Sci. 2015, 16, 20748. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.R.; Thomas, U.; Pellegrini, A. A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J. Biol. Chem. 2001, 276, 43767–43774. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, R.J.; Meisel, H. Milk protein-derived peptide inhibitors of angiotensin-I-converting enzyme. Br. J. Nutr. 2000, 84, 33–37. [Google Scholar] [CrossRef]
- Hayes, M.; Moens, L.F.; Auty, M.A.E.; Lea, T.E. Transport of a prolyl endopeptidase inhibitory peptide across the blood brain barrier demonstrated using the hCMEC/D3 cell line transcytosis assay. J. Agric. Food Chem. 2016, 64, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Usui, H.; Kurahashi, K.; Yoshikawa, M. Isolation and characterization of ovokinin, a bradykinin B1 agonist peptide derived from ovalbumin. Peptides 1995, 16, 785–790. [Google Scholar] [CrossRef]
- Miguel, M.; Aleixandre, A. Antihypertensive peptides derived from egg proteins. J. Nutr. 2006, 136, 1457–1460. [Google Scholar] [PubMed]
- Nimalaratne, C.; Bandara, N.; Wu, J. Purification and characterization of antioxidant peptides from enzymatically hydrolyzed chicken egg white. Food Chem. 2015, 188, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Kaneko, K.; Mizushige, T.; Lazarus, M.; Urade, Y.; Ohinata, K. Characterization of ovolin, an orally active tryptic peptide released from ovalbumin with anxiolytic-like activity. J. Neurochem. 2012, 122, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Voet, D.; Voet, J.G.; Pratt, C.W. Fundamentals of Biochemistry: Life at the Molecular Level, 4th ed.; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Schagger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Stryer, L. Biochemistry, 4th ed.; W. H. Freeman and Company: New York, NY, USA, 1995. [Google Scholar]
- Nicklisch, S.C.T.; Waite, J.H. Optimized DPPH assay in a detergent-based buffer system for measuring antioxidant activity of proteins. MethodsX 2014, 1, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Goupy, P.; Dufour, C.; Loonis, M.; Dangles, O. Quantitative kinetic analysis of hydrogen transfer reactions from dietary polyphenols to the DPPH radical. J. Agric. Food Chem. 2003, 51, 615–622. [Google Scholar] [CrossRef] [PubMed]


| Hydrolysate | Protein Content (%) |
|---|---|
| TRYP G + SI | 16.38 |
| LYS G | 97.89 |
| LYS G + SI | 98.48 |
| MUC G | 45.35 |
| MUC G + SI | 42.89 |
| SA G | 90.12 |
| SA G + SI | 87.71 |
| CA G | 85.56 |
| CA G + SI | 82.26 |
| Parent Protein and Hydrolysate Fraction | Peptide Amino acid Sequence | Protein Fragment | Observed Mass (Da) | Theoretical Mass (Da) | Observed m/z | Theoretical m/z | Theoretical Z |
|---|---|---|---|---|---|---|---|
| Lysozyme < 3-kDa fraction | ALLQDNIADAV | f(101–111) | 1141.60 | 1141.60 | 571.81 | 571.81 | 2 |
| ALLQDNIADAVA | f(101–112) | 1212.64 | 1212.64 | 607.33 | 607.32 | 2 | |
| ARTLKRLGMDGYRGISL | f(27–43) | 1906.06 | 1906.06 | 477.52 | 477.52 | 4 | |
| DPQGIRAWV | f(120–128) | 1040.54 | 1040.54 | 521.28 | 521.28 | 2 | |
| GIFQINSRYW a | f(73–82) | 1298.66 | 1298.64 | 650.34 | 650.33 | 2 | |
| GMDGYRGISLANWM | f(34–47) | 1569.71 | 1569.71 | 785.86 | 785.86 | 2 | |
| LGMDGYRGISL | f(33–43) | 1180.59 | 1180.59 | 591.30 | 591.30 | 2 | |
| LGMDGYRGISLA | f(33–44) | 1251.63 | 1251.63 | 626.82 | 626.82 | 2 | |
| LLQDNIADAV b | f(102–111) | 1071.54 | 1071.54 | 536.78 | 536.78 | 2 | |
| NAGDRSTDYG | f(64–73) | 1054.43 | 1054.43 | 528.22 | 528.22 | 2 | |
| NAGDRSTDYGIFQ | f(64–76) | 1442.64 | 1442.64 | 722.33 | 722.33 | 2 | |
| NAGDRSTDYGIFQI | f(64–77) | 1555.73 | 1555.73 | 778.87 | 778.87 | 2 | |
| NAGDRSTDYGIFQIN | f(64–78) | 1669.77 | 1669.77 | 835.89 | 835.89 | 2 | |
| NYNAGDRSTD | f(62–71) | 1111.45 | 1111.45 | 556.73 | 556.73 | 2 | |
| NYNAGDRSTDYGIF | f(62–75) | 1591.69 | 1591.69 | 796.85 | 796.85 | 2 | |
| NYNAGDRSTDYGIFQ | f(62–76) | 1719.76 | 1719.75 | 860.89 | 860.88 | 2 | |
| NYNAGDRSTDYGIFQI | f(62–77) | 1832.84 | 1832.83 | 917.43 | 917.42 | 2 | |
| PQGIRAWVAW | f(121–130) | 1182.63 | 1182.63 | 592.32 | 592.32 | 2 | |
| YNAGDRSTDYGIF | f(63–75) | 1477.65 | 1477.65 | 739.83 | 739.83 | 2 | |
| Lysozyme < 10-kDa fraction | CNDGKTPGAV | f(83–92) | 960.43 | 960.43 | 481.22 | 481.22 | 2 |
| CNDGKTPGAVNACHLSCS c,d | f(83–100) | 1773.73 | 1773.74 | 592.25 | 592.25 | 3 | |
| GIFQINSRYW | f(73–82) | 1282.65 | 1282.65 | 642.33 | 642.33 | 2 | |
| GMDGYRGISLANWM | f(34–47) | 1569.72 | 1569.71 | 785.87 | 785.86 | 2 | |
| NAGDRSTDYG | f(64–73) | 1054.43 | 1054.43 | 528.22 | 528.22 | 2 | |
| NAGDRSTDYGIFQI | f(64–77) | 1555.74 | 1555.73 | 778.88 | 778.87 | 2 | |
| NYNAGDRSTD | f(62–71) | 1111.45 | 1111.45 | 556.73 | 556.73 | 2 | |
| NYNAGDRSTDY | f(62–72) | 1274.52 | 1274.52 | 638.27 | 638.27 | 2 | |
| NYNAGDRSTDYG | f(62–73) | 1331.53 | 1331.54 | 666.77 | 666.78 | 2 | |
| NYNAGDRSTDYGIFQI | f(62–77) | 1832.85 | 1832.83 | 917.43 | 917.42 | 2 | |
| SALLQDNIADAV | f(100–111) | 1228.63 | 1228.63 | 615.32 | 615.32 | 2 | |
| YNAGDRSTDYG | f(63–73) | 1217.49 | 1217.49 | 609.75 | 609.75 | 2 | |
| ALLQDNIADAVACA e | f(101–114) | 1434.66 | 1434.67 | 718.34 | 718.34 | 2 | |
| Serum albumin < 3-kDa fraction | AEAKDVFLGMFL | f(344–355) | 1339.69 | 1339.68 | 1339.69 | 670.85 | 2 |
| AEFAEVSKLVTDL | f(250–262) | 1420.75 | 1420.74 | 1420.75 | 711.38 | 2 | |
| AEFAEVSKLVTDLT f | f(250–263) | 1549.79 | 1549.79 | 1549.79 | 775.90 | 2 | |
| AEFAEVSKLVTDLTK | f(250–264) | 1649.89 | 1649.89 | 1649.89 | 825.95 | 2 | |
| AEFAEVSKLVTDLTKVHT | f(250–267) | 1987.10 | 1987.06 | 1987.10 | 994.54 | 2 | |
| AEVENDEMPADLPSLA | f(315–330) | 1699.76 | 1699.76 | 1699.76 | 850.89 | 2 | |
| AEVSKLVTDLT | f(253–63) | 1174.64 | 1174.64 | 1174.64 | 588.33 | 2 | |
| AKVFDEFKPL | f(395–404) | 1192.65 | 1192.65 | 1192.65 | 597.33 | 2 | |
| AKVFDEFKPLVEEPQ | f(395–409) | 1774.91 | 1774.91 | 1774.91 | 888.46 | 2 | |
| ALEVDETYVPK | f(514–524) | 1262.64 | 1262.64 | 1262.64 | 632.33 | 2 | |
| ALEVDETYVPKE | f(514–525) | 1391.68 | 1391.68 | 1391.68 | 696.85 | 2 | |
| ALEVDETYVPKEF | f(514–526) | 1538.75 | 1538.75 | 1538.75 | 770.38 | 2 | |
| ALVLIAFA | f(45–52) | 816.51 | 816.51 | 816.51 | 409.26 | 2 | |
| ALVLIAFAQ | f(45–53) | 944.57 | 944.57 | 944.57 | 473.29 | 2 | |
| ALVLIAFAQY | f(45–54) | 1107.62 | 1107.63 | 1107.62 | 554.82 | 2 | |
| AVMDDFAAFVEK | f(570–581) | 1341.63 | 1341.63 | 1341.63 | 671.82 | 2 | |
| DDNPNLPR | f(131–138) | 939.44 | 939.44 | 939.44 | 470.73 | 2 | |
| DEFKPLVEEPQNLI | f(399–412) | 1669.86 | 1669.86 | 1669.86 | 835.94 | 2 | |
| DEFKPLVEEPQNLIK | f(399–413) | 1797.95 | 1797.95 | 1797.95 | 899.98 | 2 | |
| DETYVPKE | f(518–525) | 979.45 | 979.45 | 979.45 | 490.73 | 2 | |
| DLGEENFKALV | f(37–47) | 1233.63 | 1233.62 | 1233.63 | 617.82 | 2 | |
| DLGEENFKALVL | f(37–48) | 1346.71 | 1346.71 | 1346.71 | 674.36 | 2 | |
| DLPSLAADFVES | f(325–336) | 1262.60 | 1262.60 | 1262.60 | 632.31 | 2 | |
| DLPSLAADFVESK | f(325–337) | 1390.70 | 1390.70 | 1390.70 | 696.36 | 2 | |
| DVFLGMFLYE g | f(348–357) | 1248.77 | 1248.57 | 1248.77 | 625.29 | 2 | |
| EDHVKLVNEVTEFA | f(61–74) | 1628.81 | 1628.80 | 1628.81 | 815.41 | 2 | |
| ENDEMPADLPSL | f(318–329) | 1329.58 | 1329.58 | 1329.58 | 665.80 | 2 | |
| ENDEMPADLPSLAADFVES | f(318–336) | 2048.90 | 2048.89 | 2048.90 | 1025.45 | 2 | |
| ENDEMPADLPSLAADFVESK | f(318–337) | 2177.01 | 2176.98 | 2177.01 | 1089.50 | 2 | |
| EQLKAVMDD | f(566–574) | 1047.49 | 1047.49 | 1047.49 | 524.75 | 2 | |
| EQLKAVMDDF h | f(566–575) | 1176.55 | 1176.55 | 1176.55 | 589.28 | 2 | |
| EQLKAVMDDFA h | f(566–576) | 1247.59 | 1247.59 | 1247.59 | 624.80 | 2 | |
| EQLKAVMDDFAA h | f(566–577) | 1318.62 | 1318.62 | 1318.62 | 660.32 | 2 | |
| EVDETYVPK | f(516–524) | 1078.52 | 1078.52 | 1078.52 | 540.27 | 2 | |
| EVDETYVPKE | f(516–525) | 1207.57 | 1207.56 | 1207.57 | 604.79 | 2 | |
| EVDETYVPKEF | f(516–526) | 1354.63 | 1354.63 | 1354.63 | 678.32 | 2 | |
| EVDETYVPKEFN | f(516–527) | 1468.67 | 1468.67 | 1468.67 | 735.34 | 2 | |
| EVDETYVPKEFNAET i | f(516–530) | 1770.79 | 1770.78 | 1770.79 | 886.40 | 2 | |
| FDEFKPLVEEPQNLIK | f(398–413) | 1945.02 | 1945.02 | 1945.02 | 973.52 | 2 | |
| FKDLGEEN | f(35–42) | 950.44 | 950.43 | 950.44 | 476.22 | 2 | |
| FKDLGEENFKALV | f(35–47) | 1508.79 | 1508.79 | 1508.79 | 755.40 | 2 | |
| FKDLGEENFKALVL | f(35–48) | 1621.87 | 1621.87 | 1621.87 | 811.94 | 2 | |
| FPKAEFAEVSKLVTDLT | f(247–263) | 1894.01 | 1894.01 | 1894.01 | 632.34 | 3 | |
| FYAPELLFFAK | f(173–183) | 1344.71 | 1344.71 | 1344.71 | 673.36 | 2 | |
| KPLVEEPQN | f(402–410) | 1052.55 | 1052.55 | 1052.55 | 527.28 | 2 | |
| KPLVEEPQNLI | f(402–412) | 1278.72 | 1278.72 | 1278.72 | 640.37 | 2 | |
| KVPQVSTPT | f(438–446) | 955.53 | 955.53 | 955.53 | 478.77 | 2 | |
| KVPQVSTPTLV | f(438–448) | 1167.69 | 1167.69 | 1167.69 | 584.85 | 2 | |
| KVPQVSTPTLVEV | f(438–450) | 1395.79 | 1395.80 | 1395.79 | 698.91 | 2 | |
| KVPQVSTPTLVEVS | f(438–451) | 1482.83 | 1482.83 | 1482.83 | 742.42 | 2 | |
| LDELRDEG | f(206–213) | 945.44 | 945.44 | 945.44 | 473.73 | 2 | |
| LEVDETYVPK | f(515–524) | 1191.60 | 1191.60 | 1191.60 | 596.81 | 2 | |
| LEVDETYVPKE | f(515–525) | 1320.65 | 1320.64 | 1320.65 | 661.33 | 2 | |
| LQHKDDNPNLPR | f(127–138) | 1445.73 | 1445.74 | 1445.73 | 482.92 | 3 | |
| LVAASQAALG | f(599–608) | 899.51 | 899.51 | 899.51 | 450.76 | 2 | |
| LVNEVTEFA | f(66–74) | 1020.52 | 1020.51 | 1020.52 | 511.26 | 2 | |
| LVNEVTEFAK | f(66–75) | 1148.61 | 1148.61 | 1148.61 | 575.31 | 2 | |
| LVNEVTEFAKTCVADESAENCDK c,d | f(66–88) | 2512.11 | 2512.13 | 2512.11 | 629.04 | 4 | |
| LVRPEVDVM g | f(139–147) | 1072.56 | 1072.56 | 1072.56 | 537.29 | 2 | |
| NDEMPADLPSLA i | f(318–330) | 1272.56 | 1272.55 | 1272.56 | 637.28 | 2 | |
| NDEMPADLPSLAADF | f(318–333) | 1604.71 | 1604.70 | 1604.71 | 803.36 | 2 | |
| NDEMPADLPSLAADFVES | f(318–336) | 1919.86 | 1919.85 | 1919.86 | 960.93 | 2 | |
| NDEMPADLPSLAADFVESK | f(318–337) | 2047.95 | 2047.94 | 2047.95 | 1024.98 | 2 | |
| NEVTEFAKTCVADESAENCDK c,d | f(68–88) | 2299.96 | 2299.98 | 2299.96 | 1150.99 | 2 | |
| NRRPCFSALEVDETYVPKE d,j | f(507–525) | 2199.96 | 2200.09 | 2199.96 | 734.37 | 3 | |
| NYAEAKDVFLG | f(342–352) | 1225.60 | 1225.60 | 1225.60 | 613.81 | 2 | |
| NYAEAKDVFLGM | f(342–353) | 1356.64 | 1356.64 | 1356.64 | 679.33 | 2 | |
| NYAEAKDVFLGMFL | f(342–355) | 1616.83 | 1616.79 | 1616.83 | 809.40 | 2 | |
| QHKDDNPNLPR h | f(128–138) | 1315.62 | 1315.63 | 1315.62 | 439.55 | 3 | |
| QYLQQCPFEDHV d | f(53–64) | 1471.68 | 1471.67 | 1471.68 | 736.84 | 2 | |
| RETYGEM | f(105–111) | 884.37 | 884.37 | 884.37 | 443.19 | 2 | |
| RHPDYSVV | f(361–368) | 971.48 | 971.48 | 971.48 | 486.75 | 2 | |
| RHPDYSVVL | f(361–369) | 1084.57 | 1084.57 | 1084.57 | 543.29 | 2 | |
| SALEVDETYVPK | f(513–524) | 1349.67 | 1349.67 | 1349.67 | 675.84 | 2 | |
| SALEVDETYVPKE | f(513–525) | 1478.72 | 1478.71 | 1478.72 | 740.36 | 2 | |
| SALEVDETYVPKEF | f(513–526) | 1625.79 | 1625.78 | 1625.79 | 813.90 | 2 | |
| TPVSDRVT | f(491–498) | 873.45 | 873.46 | 873.45 | 437.74 | 2 | |
| VFDEFKP | f(397–403) | 880.43 | 880.43 | 880.43 | 441.22 | 2 | |
| VFDEFKPL | f(397–404) | 993.52 | 993.52 | 993.52 | 497.77 | 2 | |
| VFDEFKPLVEEPQ | f(397–409) | 1575.78 | 1575.78 | 1575.78 | 788.90 | 2 | |
| VFDEFKPLVEEPQN | f(397–410) | 1689.83 | 1689.82 | 1689.83 | 845.92 | 2 | |
| VFDEFKPLVEEPQNLI | f(397–412) | 1916.00 | 1915.99 | 1916.00 | 959.00 | 2 | |
| VKLVNEVTEFA | f(64–74) | 1247.68 | 1247.68 | 1247.68 | 624.85 | 2 | |
| VLIAFAQYL | f(47–55) | 1036.60 | 1036.60 | 1036.60 | 519.31 | 2 | |
| VPQVSTPTLVEVS | f(439–451) | 1354.74 | 1354.73 | 1354.74 | 678.37 | 2 | |
| VSTPTLVEVS | f(442–451) | 1030.56 | 1030.55 | 1030.56 | 516.28 | 2 | |
| DVFLGMFL g | f(348–355) | 956.47 | 956.47 | 956.47 | 479.24 | 2 | |
| TLRETYGEMADCCAKQEPERNECFLQHKDDNPNLPR d,i | f(103–138) | 4216.84 | 4216.90 | 4216.84 | 844.39 | 5 | |
| LVNEVTEFAKTC d | f(66–77) | 1318.71 | 1318.68 | 1318.71 | 660.35 | 2 | |
| ENDEMPADLPSLA h | f(318–330) | 1382.60 | 1382.60 | 1382.60 | 692.31 | 2 | |
| LSVVLNQLCVL c | f(477–87) | 1231.69 | 1231.68 | 1231.69 | 616.85 | 2 | |
| Serum albumin < 10-kDa fraction | AEAKDVFLGMF | f(344–354) | 1226.60 | 1226.60 | 614.31 | 614.31 | 2 |
| AEAKDVFLGMFL | f(344–355) | 1339.70 | 1339.68 | 670.86 | 670.85 | 2 | |
| AEFAEVSKLVTDL | f(250–262) | 1420.76 | 1420.74 | 711.39 | 711.38 | 2 | |
| AEFAEVSKLVTDLT | f(250–263) | 1521.91 | 1521.79 | 761.96 | 761.90 | 2 | |
| AEFAEVSKLVTDLTK | f(250–264) | 1649.88 | 1649.89 | 550.97 | 550.97 | 3 | |
| AEVSKLVTDLT | f(253–263) | 1174.65 | 1174.64 | 588.33 | 588.33 | 2 | |
| AKVFDEFKPLVEEPQNLIK | f(395–413) | 2243.25 | 2243.22 | 748.76 | 748.75 | 3 | |
| ALVLIAFAQ | f(45–53) | 944.56 | 944.57 | 473.29 | 473.29 | 2 | |
| ALVLIAFAQY | f(45–54) | 1107.63 | 1107.63 | 554.82 | 554.82 | 2 | |
| APELLFFAK | f(175–183) | 1034.57 | 1034.58 | 518.29 | 518.30 | 2 | |
| AVMDDFAAFVE | f(570–580) | 1213.54 | 1213.53 | 607.78 | 607.77 | 2 | |
| AVMDDFAAFVEK | f(570–581) | 1341.64 | 1341.63 | 671.83 | 671.82 | 2 | |
| DEFKPLVEEPQNL | f(399–411) | 1556.80 | 1556.77 | 779.41 | 779.39 | 2 | |
| DEFKPLVEEPQNLI | f(399–412) | 1669.89 | 1669.86 | 835.95 | 835.94 | 2 | |
| DEFKPLVEEPQNLIK | f(399–413) | 1798.00 | 1797.95 | 900.01 | 899.98 | 2 | |
| DLGEENFKALVLI | f(37–49) | 1459.82 | 1459.79 | 730.92 | 730.90 | 2 | |
| DLGEENFKALVLIA | f(37–50) | 1530.85 | 1530.83 | 766.43 | 766.42 | 2 | |
| DLGEENFKALVLIAFAQ | f(37–53) | 1877.06 | 1876.99 | 939.54 | 939.50 | 2 | |
| DLPSLAADF | f(325–333) | 947.45 | 947.46 | 474.73 | 474.74 | 2 | |
| DLPSLAADFVES | f(325–334) | 1262.61 | 1262.60 | 632.31 | 632.31 | 2 | |
| DLPSLAADFVESK | f(325–335) | 1390.72 | 1390.70 | 696.37 | 696.36 | 2 | |
| DVFLGMFLY g | f(348–366) | 1119.53 | 1119.53 | 560.77 | 560.77 | 2 | |
| DVFLGMFLYE g | f(348–367) | 1248.57 | 1248.57 | 625.29 | 625.29 | 2 | |
| DVFLGMFLYEYA g | f(348–369) | 1482.70 | 1482.67 | 742.36 | 742.34 | 2 | |
| ENDEMPADLPSLA | f(318–330) | 1400.63 | 1400.61 | 701.32 | 701.31 | 2 | |
| EQLKAVMDDF | f(566–575) | 1176.55 | 1176.55 | 589.28 | 589.28 | 2 | |
| EQLKAVMDDFAA | f(566–577) | 1318.63 | 1318.62 | 660.32 | 660.32 | 2 | |
| EQLKAVMDDFAAF | f(566–578) | 1483.73 | 1483.70 | 742.87 | 742.86 | 2 | |
| EVDETYVPKE | f(516–525) | 1207.56 | 1207.56 | 604.79 | 604.79 | 2 | |
| FDEFKPLVEEPQNLIK | f(398–413) | 1945.04 | 1945.02 | 649.36 | 649.35 | 3 | |
| FKDLGEENFKALV | f(35–47) | 1508.81 | 1508.79 | 755.41 | 755.40 | 2 | |
| FKDLGEENFKALVL | f(35–48) | 1621.91 | 1621.87 | 811.96 | 811.94 | 2 | |
| FKDLGEENFKALVLI | f(35–49) | 1735.00 | 1734.96 | 868.51 | 868.49 | 2 | |
| FKDLGEENFKALVLIA | f(35–50) | 1806.04 | 1805.99 | 904.03 | 904.00 | 2 | |
| FKDLGEENFKALVLIAFAQ | f(35–53) | 2152.24 | 2152.16 | 1077.13 | 1077.09 | 2 | |
| FPKAEFAEVSKLVTDLT | f(247–263) | 1894.01 | 1894.01 | 632.35 | 632.34 | 3 | |
| FYAPELLFFA | f(173–182) | 1216.62 | 1216.62 | 609.32 | 609.32 | 2 | |
| FYAPELLFFAK | f(173–183) | 1344.73 | 1344.71 | 673.37 | 673.36 | 2 | |
| FYAPELLFFAKR | f(173–184) | 1500.79 | 1500.81 | 501.27 | 501.28 | 3 | |
| KDLGEENFKALVLIAFAQ | f(36–53) | 2005.11 | 2005.09 | 669.38 | 669.37 | 3 | |
| KVPQVSTPT | f(438–446) | 955.52 | 955.53 | 478.77 | 478.77 | 2 | |
| KVPQVSTPTLVEV | f(438–450) | 1395.81 | 1395.80 | 698.91 | 698.91 | 2 | |
| KVPQVSTPTLVEVS | f(438–451) | 1482.85 | 1482.83 | 742.43 | 742.42 | 2 | |
| KVPQVSTPTLVEVSRN k | f(438–453) | 1710.00 | 1709.92 | 856.01 | 855.97 | 2 | |
| LQHKDDNPNLPR | f(127–138) | 1445.71 | 1445.74 | 482.91 | 482.92 | 3 | |
| LVNEVTEFAKTCVADESAENCDKSLHTLF c,k | f(66–94) | 3210.60 | 3210.50 | 1071.21 | 1071.17 | 3 | |
| NDEMPADLPSLA | f(319–330) | 1271.58 | 1271.57 | 636.80 | 636.79 | 2 | |
| NDEMPADLPSLAADFVESK | f(319–337) | 2048.01 | 2047.94 | 1025.01 | 1024.98 | 2 | |
| NYAEAKDVFLGMF | f(342–354) | 1503.73 | 1503.71 | 752.87 | 752.86 | 2 | |
| NYAEAKDVFLGMFL g | f(342–355) | 1632.81 | 1632.79 | 817.41 | 817.40 | 2 | |
| RHPDYSVVLLL | f(361–371) | 1310.74 | 1310.73 | 656.38 | 656.37 | 2 | |
| SALEVDETYVPK | f(513–524) | 1349.68 | 1349.67 | 675.85 | 675.84 | 2 | |
| TKKVPQVSTPTLVEVSf | f(436–451) | 1739.99 | 1739.97 | 871.00 | 870.99 | 2 | |
| TYETTLEK | f(376–383) | 983.47 | 983.48 | 492.74 | 492.75 | 2 | |
| VFDEFKPL | f(397–404) | 993.50 | 993.52 | 497.76 | 497.77 | 2 | |
| VFDEFKPLVEEPQ | f(397–409) | 1575.81 | 1575.78 | 788.91 | 788.90 | 2 | |
| VFDEFKPLVEEPQN | f(397–410) | 1689.86 | 1689.82 | 845.94 | 845.92 | 2 | |
| VFDEFKPLVEEPQNL | f(397–411) | 1802.96 | 1802.91 | 902.49 | 902.46 | 2 | |
| VFDEFKPLVEEPQNLI | f(397–412) | 1916.05 | 1915.99 | 959.03 | 959.00 | 2 | |
| VFDEFKPLVEEPQNLIK | f(397–413) | 2044.11 | 2044.09 | 682.38 | 682.37 | 3 | |
| VFDEFKPLVEEPQNLIKQ | f(397–414) | 2172.21 | 2172.15 | 725.08 | 725.06 | 3 | |
| VLIAFAQYL | f(47–55) | 1036.58 | 1036.60 | 519.30 | 519.31 | 2 | |
| VPQVSTPTLVEV | f(439–450) | 1267.71 | 1267.70 | 634.86 | 634.86 | 2 | |
| VPQVSTPTLVEVS | f(439–451) | 1354.75 | 1354.73 | 678.38 | 678.37 | 2 | |
| EQLKAVMDDFA | f(566–576) | 1265.60 | 1265.60 | 633.81 | 633.81 | 2 | |
| YAPELLFFAK | f(174–183) | 1197.65 | 1197.64 | 599.83 | 599.83 | 2 | |
| LVRPEVDV | f(139–146) | 925.51 | 925.52 | 463.76 | 463.77 | 2 | |
| EVDETYVPK | f(516–524) | 1078.51 | 1078.52 | 540.26 | 540.27 | 2 | |
| LSQRFPKAEFAEVSKLVTDLT | f(243–263) | 2379.28 | 2378.28 | 595.83 | 595.58 | 4 | |
| VLIAFAQY | f(47–54) | 923.49 | 923.51 | 462.75 | 462.76 | 2 |
| Protein | Chain Length | Molecular Weight (Mw) (KDa) | Uniprot Accession No. |
|---|---|---|---|
| Gut cryptome protein | – | – | – |
| Trypsin (TRYP) | 223 | 24 | P00761 |
| Lysozyme (LYS) | 148 | 17 | P61626 |
| Porcine salivary mucin (Apomucin) (MUC) | 1150 | 110 | P12021 |
| Serum albumin (SA) | 609 | 69 | P02768 |
| Dietary protein | – | – | – |
| Chicken albumin (CA) | 385 | 43 | P01012 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dave, L.A.; Hayes, M.; Mora, L.; Montoya, C.A.; Moughan, P.J.; Rutherfurd, S.M. Gastrointestinal Endogenous Protein-Derived Bioactive Peptides: An in Vitro Study of Their Gut Modulatory Potential. Int. J. Mol. Sci. 2016, 17, 482. https://doi.org/10.3390/ijms17040482
Dave LA, Hayes M, Mora L, Montoya CA, Moughan PJ, Rutherfurd SM. Gastrointestinal Endogenous Protein-Derived Bioactive Peptides: An in Vitro Study of Their Gut Modulatory Potential. International Journal of Molecular Sciences. 2016; 17(4):482. https://doi.org/10.3390/ijms17040482
Chicago/Turabian StyleDave, Lakshmi A., Maria Hayes, Leticia Mora, Carlos A. Montoya, Paul J. Moughan, and Shane M. Rutherfurd. 2016. "Gastrointestinal Endogenous Protein-Derived Bioactive Peptides: An in Vitro Study of Their Gut Modulatory Potential" International Journal of Molecular Sciences 17, no. 4: 482. https://doi.org/10.3390/ijms17040482