Molecular Characterization of MaCCS, a Novel Copper Chaperone Gene Involved in Abiotic and Hormonal Stress Responses in Musa acuminata cv. Tianbaojiao
Abstract
:1. Introduction
2. Results
2.1. Identification of the CCS Gene in Banana
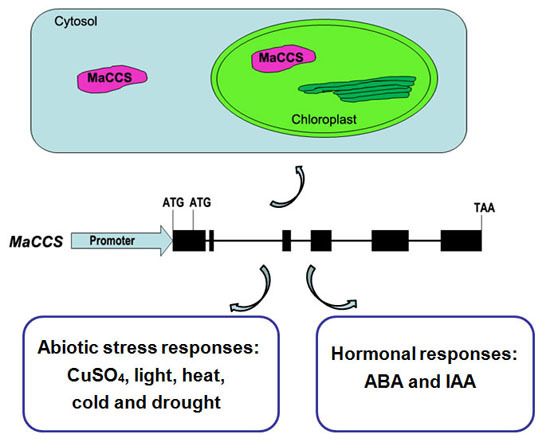
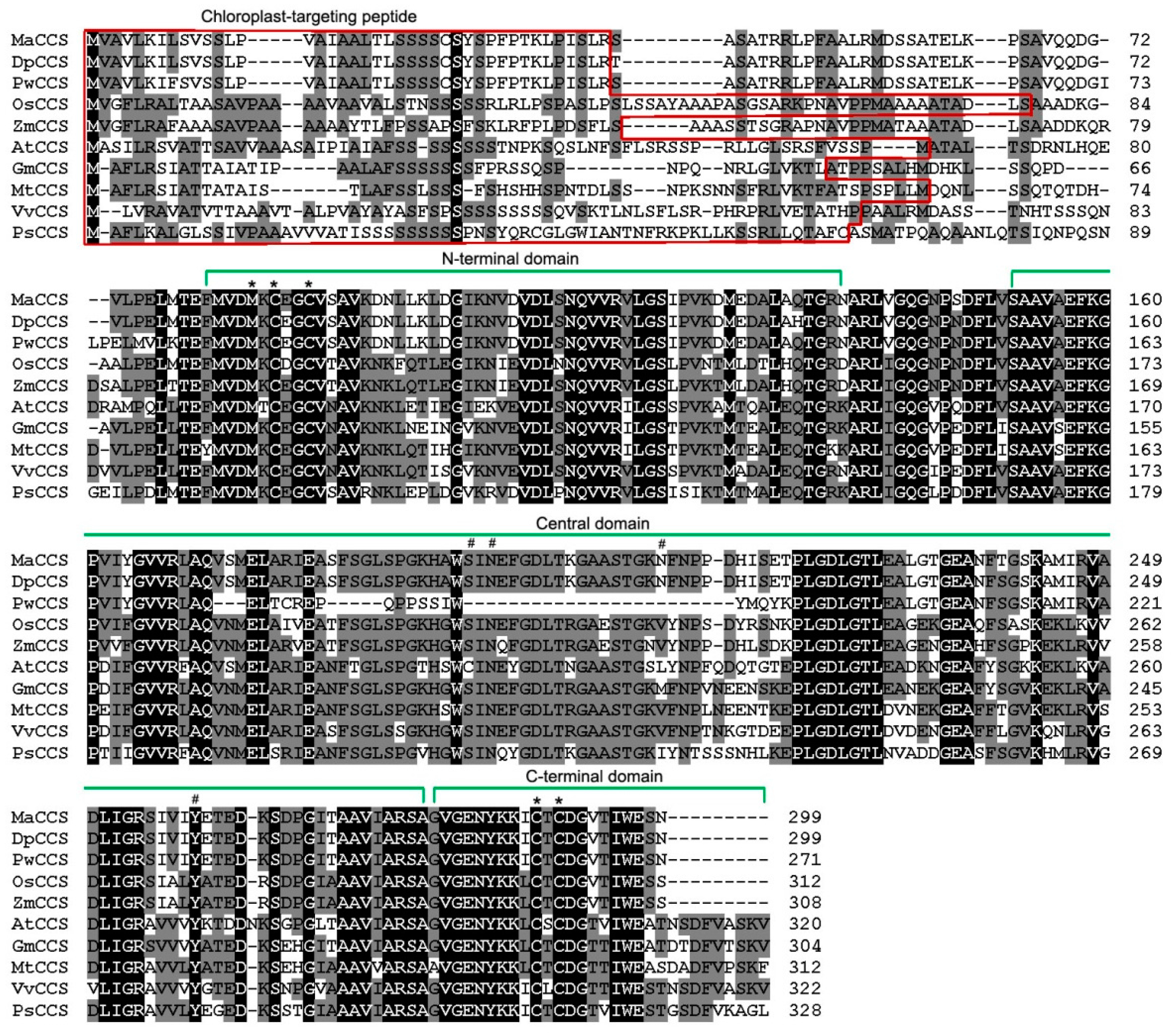
2.2. Gene Structure and Phylogenetic Analysis of MaCCS
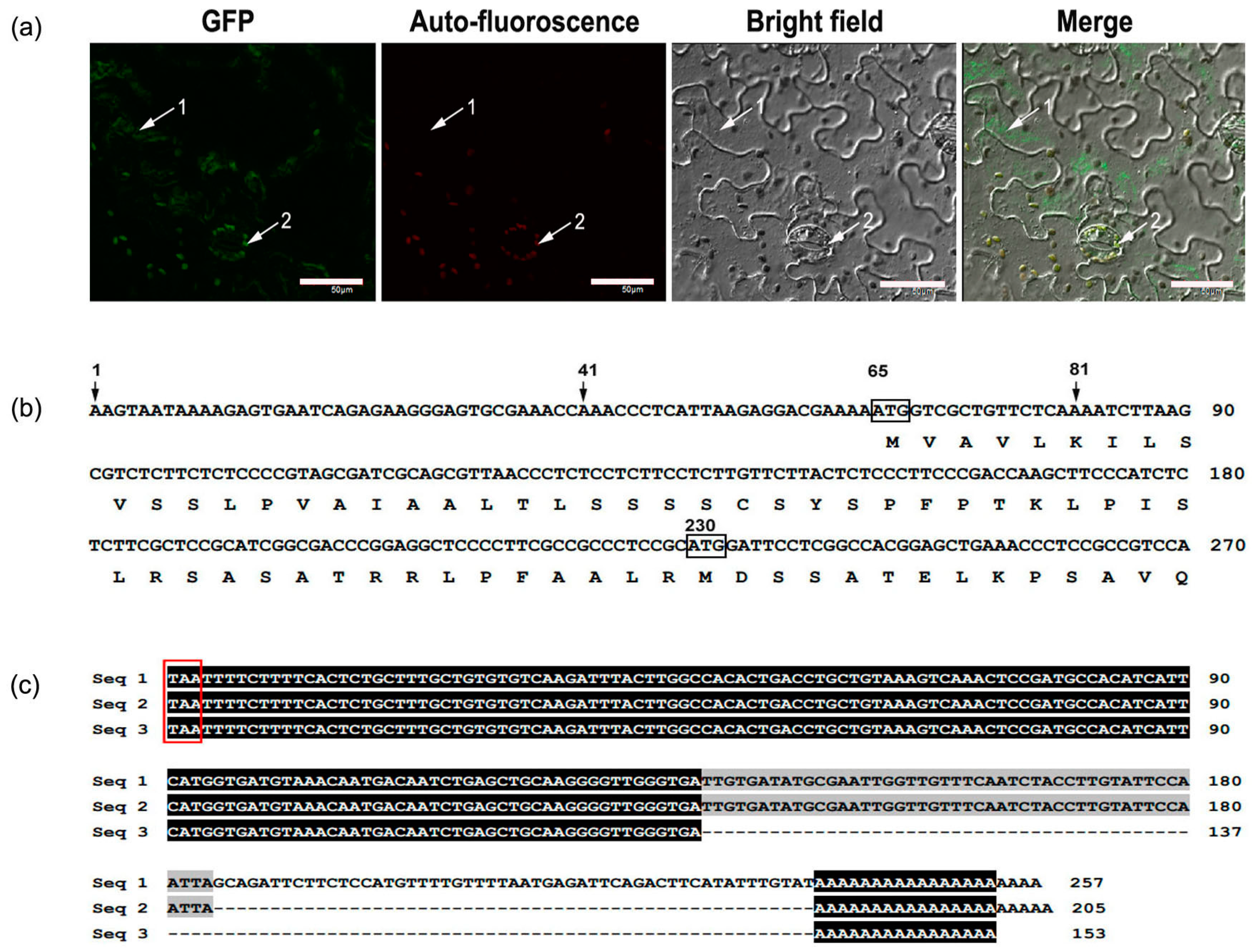
2.3. Subcellular Localization of MaCCS
2.4. Analysis of the Transcription Start Site and 3′ Untranslated Region of MaCCS
2.5. MaCCS Promoter Isolation and Cis-Element Analysis
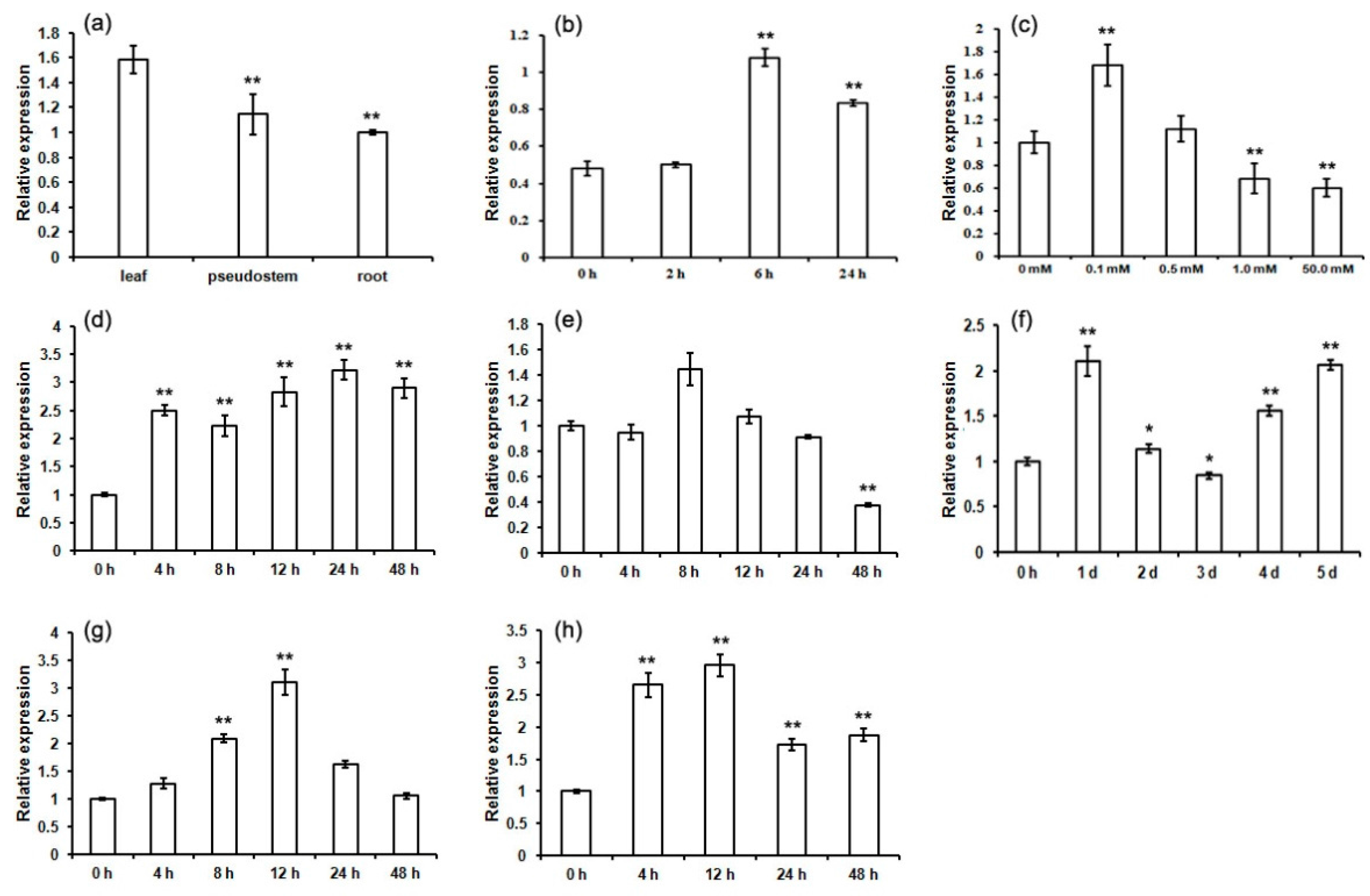
2.6. Expression Patterns of MaCCS under Abiotic and Hormonal Stresses
3. Discussion
3.1. A Single MaCCS Gene with Diverse Transcripts
3.2. The Differential Expression of MaCCS in Different Tissues
3.3. MaCCS Is Involved in Abiotic and Hormonal Stress Responses
4. Materials and Methods
4.1. Plant Material and Stress Treatments
4.2. Sequence Retrieval and Gene Cloning
4.3. Sequence Analysis
4.4. Subcellular Localization Analysis
4.5. Expression Analysis by Quantitative Real-Time PCR
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Cu/ZnSOD | Copper/Zinc superoxide dismutase |
| CCS | Copper chaperone for superoxide dismutase |
| ORF | Open reading frame |
| ATSSs | Alternative transcription start sites |
| qRT-PCR | Quantitative real-time PCR |
| PKW | Pisang Klutuk Wulang |
| ABA | Abscisic acid |
| IAA | Indole-3-acietic acid |
| NCBI | National Center for Biotechnology Information Search database |
| ATG | The start codon |
| GFP | The green fluorescent protein |
| 5′ UTR | 5′ untranslated region |
| 3′ UTR | 3′ untranslated region |
| MaCAC | Clathrin adaptor complexes medium |
References
- Karuppanapandian, T.; Moon, J.C.; Kim, C.; Manoharan, K.; Kim, W. Reactive oxygen species in plants: Their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop Sci. 2011, 5, 709–725. [Google Scholar]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Fridovich, I. On the stability of bovine superoxide dismutase: The effects of metals. J. Biol. Chem. 1973, 248, 2645–2649. [Google Scholar] [PubMed]
- Rae, T.; Schmidt, P.; Pufahl, R.; Culotta, V.; O'halloran, T. Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase. Science 1999, 284, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.J.; Winge, D.R. Copper metallochaperones. Annu. Revi. Biochem. 2010, 79, 537–562. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.C.; Lee, W.C.; Guo, W.Y.; Pan, S.M.; Chen, L.J.; Li, H.M.; Jinn, T.L. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol. 2005, 139, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Shipp, E.; Sanchez, R.J.; Liba, A.; Stine, J.E.; Hart, P.J.; Gralla, E.B.; Nersissian, A.M.; Valentine, J.S. Cobalt(2+) binding to human and tomato copper chaperone for superoxide dismutase: Implications for the metal ion transfer mechanism. Biochemistry 2000, 39, 5413–5421. [Google Scholar] [CrossRef] [PubMed]
- Trindade, L.M.; Horvath, B.M.; Bergervoet, M.J.E.; Visser, R.G.F. Isolation of a gene encoding a copper chaperone for the copper/zinc superoxide dismutase and characterization of its promoter in potato. Plant Physiol. 2003, 133, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Lai, Z.X. Cloning of copper chaperone for superoxide dismutase gene CCS from embryogenic dallus of Dimocarpus longan Lour. and its expression analysis during somatic embryogenesis. Chin. J. Appl. Environ. Biol. 2012, 18, 351–358. [Google Scholar] [CrossRef]
- Molina-Rueda, J.J.; Tsai, C.J.; Kirby, E.G. The populus superoxide dismutase gene family and its responses to drought stress in transgenic poplar overexpressing a pine cytosolic glutamine synthetase (GS1a). PLOS ONE 2013, 8, e56421. [Google Scholar] [CrossRef] [PubMed]
- Sagasti, S.; Bernal, M.; Sancho, D.; Castillo, M.B.; Picorel, R. Regulation of the chloroplastic copper chaperone (CCS) and cuprozinc superoxide dismutase (CSD2) by alternative splicing and copper excess in Glycine max. Funct. Plant Biol. 2014, 41, 144–155. [Google Scholar] [CrossRef]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genom. 2015, 16, 823. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Hou, P.; Lu, Y.; Deng, S.; Li, N.; Zhao, R.; Sun, J.; Wang, Y.; Han, Y.; Lang, T.; et al. Overexpression of copper/zinc superoxide dismutase from mangrove Kandelia candel in tobacco enhances salinity tolerance by the reduction of reactive oxygen species in chloroplast. Front. Plant Sci. 2015, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Negi, N.P.; Shrivastava, D.C.; Sharma, V.; Sarin, N.B. Overexpression of Cu/ZnSOD from Arachis hypogaea alleviates salinity and drought stress in tobacco. Plant Cell Rep. 2015, 34, 1109–1126. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davey, M.W.; Gudimella, R.; Harikrishna, J.A.; Sin, L.W.; Khalid, N.; Keulemans, J. A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genom. 2013, 14, 683. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, R.; Kageyama, T. Structure, gene expression, and evolution of primate copper chaperone for superoxide dismutase. Gene 2013, 516, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.J.; Rae, T.D.; Pufahl, R.A.; Hamma, T.; Strain, J.; O'Halloran, T.V.; Culotta, V.C. Multiple protein domains contribute to the action of the copper chaperone for superoxide dismutase. J. Biol. Chem. 1999, 274, 23719–23725. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.J.; Kunst, C.; Culotta, V.C. Copper activation of superoxide dismutase 1 (SOD1) in vivo: Role for protein-protein interactions with the copper chaperone for SOD1. J. Biol. Chem. 2000, 275, 33771–33776. [Google Scholar] [CrossRef] [PubMed]
- Lamb, A.L.; Torres, A.S.; O'Halloran, T.V.; Rosenzweig, A.C. Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 2001, 8, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Menkens, A.E.; Schindler, U.; Cashmore, A.R. The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 1995, 20, 506–510. [Google Scholar] [CrossRef]
- Lin, Y.L.; Lai, Z.X. Superoxide dismutase multigene family in longan somatic embryos: A comparison of CuZn-SOD, Fe-SOD, and Mn-SOD gene structure, splicing, phylogeny, and expression. Mol. Breed. 2013, 32, 595–615. [Google Scholar] [CrossRef]
- Hartmann, U.; Valentine, W.J.; Christie, J.M.; Hays, J.; Jenkins, G.I.; Weisshaar, B. Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol. Biol. 1998, 36, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol. 1993, 101, 1119–1120. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.; Hagen, G.; Li, Y.; Ulmasov, T.; Liu, Z.; Strabala, T.; Gee, M. Auxin-regulated transcription. Funct. Plant Biol. 1993, 20, 489–502. [Google Scholar] [CrossRef]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [PubMed]
- Klotz, K.L.; Lagrimini, L.M. Phytohormone control of the tobacco anionic peroxidase promoter. Plant Mol. Biol. 1996, 31, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Freitas, F.Z.; Bertolini, M.C. Genomic organization of the Neurospora crassa gsn gene: Possible involvement of the STRE and HSE elements in the modulation of transcription during heat shock. Mol. Genet. Genom. 2004, 272, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Diaz-De-Leon, F.; Klotz, K.L.; Lagrimini, L.M. Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxidase gene. Plant Physiol. 1993, 101, 1117–1118. [Google Scholar] [CrossRef] [PubMed]
- Daraselia, N.D.; Tarchevskaya, S.; Narita, J.O. The promoter for tomato 3-hydroxy-3-methylglutaryl coenzyme A reductase gene 2 has unusual regulatory elements that direct high-level expression. Plant Physiol. 1996, 112, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, N.W.; Shepherd, K. The taxonomy and origins of the cultivated bananas. J. Linn. Soc. Lond. Bot. 1955, 55, 302–312. [Google Scholar] [CrossRef]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Gao, J.; He, W.; Dou, T.; Ding, L.; Wu, J.; Li, C.; Peng, X.; Zhang, S.; Yi, G. Comparative transcriptomics analysis reveals difference of key gene expression between banana and plantain in response to cold stress. BMC Genom. 2015, 16, 446. [Google Scholar] [CrossRef] [PubMed]
- Beauclair, L.; Yu, A.; Bouche, N. microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 2010, 62, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.X.; Lai, Z.X.; He, S.L.; Huang, Y.J.; Lin, Y.L.; Wu, J.S. Development of a transgenic acceptor system for banana. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 2006, 35, 509–514. [Google Scholar]
- Droc, G.; Lariviere, D.; Guignon, V.; Yahiaoui, N.; This, D.; Garsmeur, O.; Dereeper, A.; Hamelin, C.; Argout, X.; Dufayard, J.F.; et al. The Banana Genome Hub. Database 2013. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2013. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Heijne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.R.R.; Kirti, P.B. A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiol. Biochem. 2010, 48, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, H.Y.; Kuang, J.F.; Li, J.G.; Lu, W.J.; Chen, J.Y. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Chen, F.; Liu, W.; Thu, M.K.; Zhang, Z.; Chen, Y.; Cheng, C.; Lin, Y.; Wang, T.; Lai, Z. Molecular Characterization of MaCCS, a Novel Copper Chaperone Gene Involved in Abiotic and Hormonal Stress Responses in Musa acuminata cv. Tianbaojiao. Int. J. Mol. Sci. 2016, 17, 441. https://doi.org/10.3390/ijms17040441
Feng X, Chen F, Liu W, Thu MK, Zhang Z, Chen Y, Cheng C, Lin Y, Wang T, Lai Z. Molecular Characterization of MaCCS, a Novel Copper Chaperone Gene Involved in Abiotic and Hormonal Stress Responses in Musa acuminata cv. Tianbaojiao. International Journal of Molecular Sciences. 2016; 17(4):441. https://doi.org/10.3390/ijms17040441
Chicago/Turabian StyleFeng, Xin, Fanglan Chen, Weihua Liu, Min Kyaw Thu, Zihao Zhang, Yukun Chen, Chunzhen Cheng, Yuling Lin, Tianchi Wang, and Zhongxiong Lai. 2016. "Molecular Characterization of MaCCS, a Novel Copper Chaperone Gene Involved in Abiotic and Hormonal Stress Responses in Musa acuminata cv. Tianbaojiao" International Journal of Molecular Sciences 17, no. 4: 441. https://doi.org/10.3390/ijms17040441