Starvation after Cobalt-60 γ-Ray Radiation Enhances Metastasis in U251 Glioma Cells by Regulating the Transcription Factor SP1
Abstract
:1. Introduction
2. Results
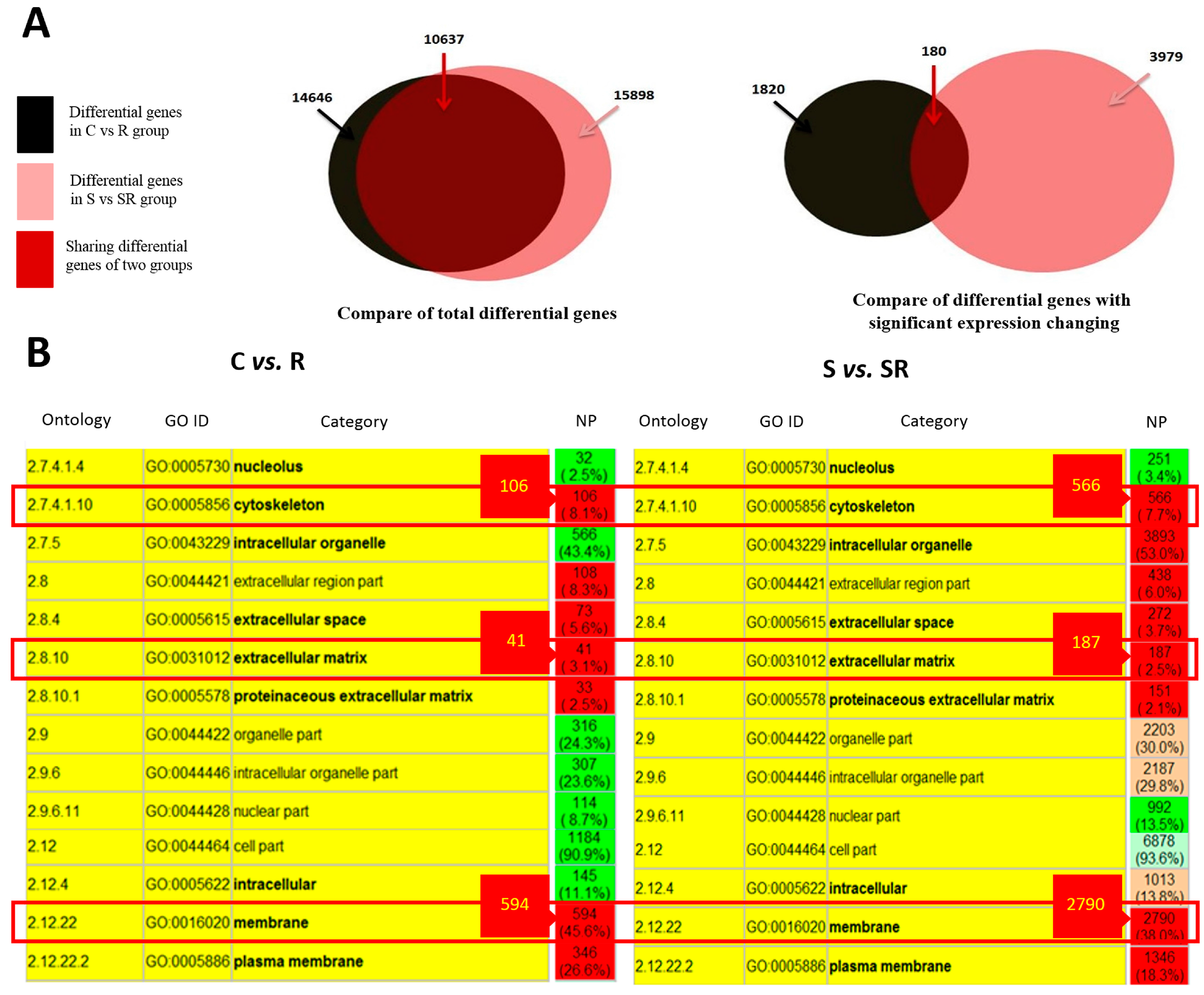
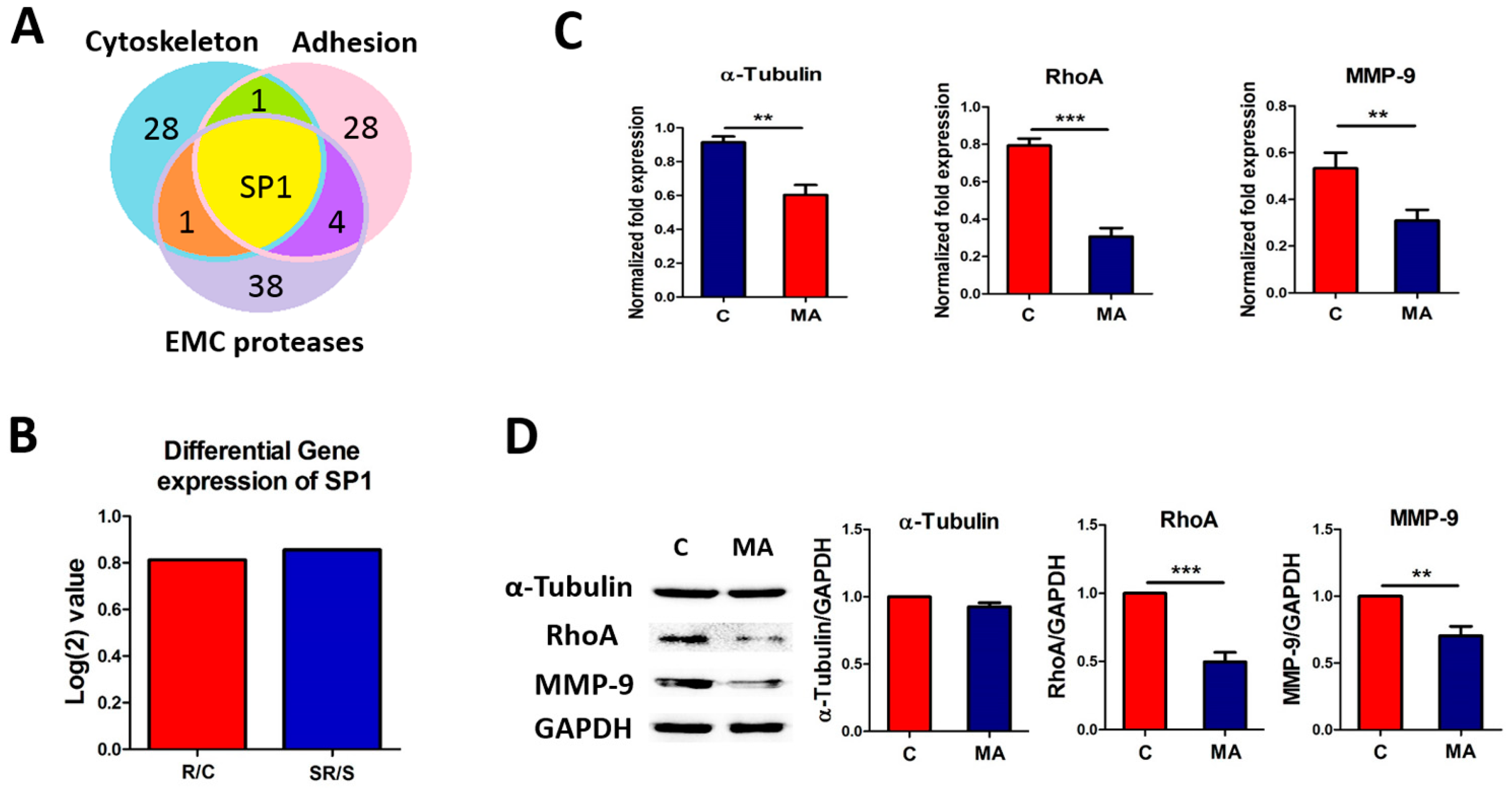
2.1. Transcriptome Analysis Demonstrated Distinct Expression Profiles Following Radiation and Starvation Treatments
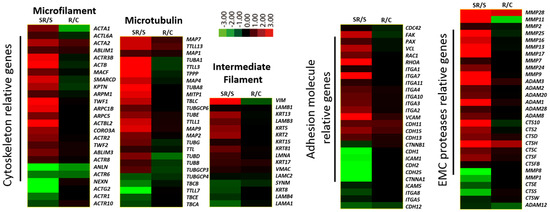
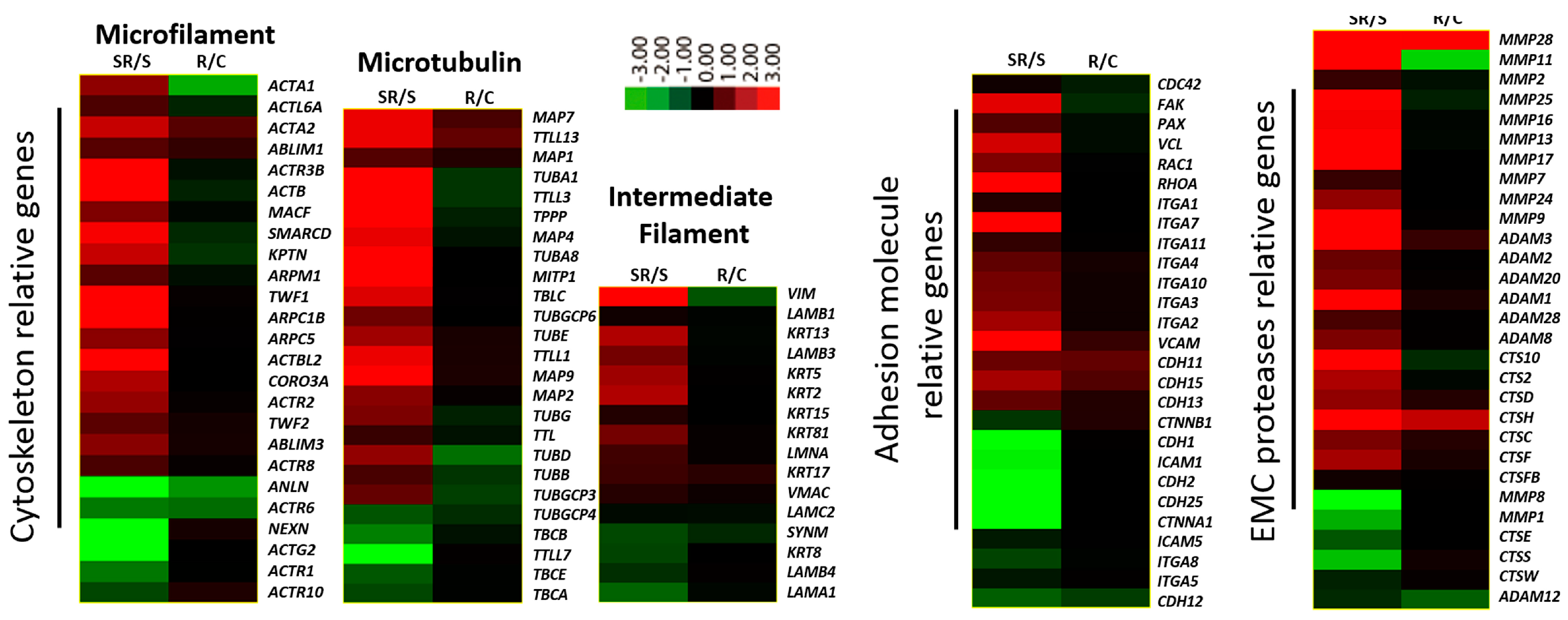
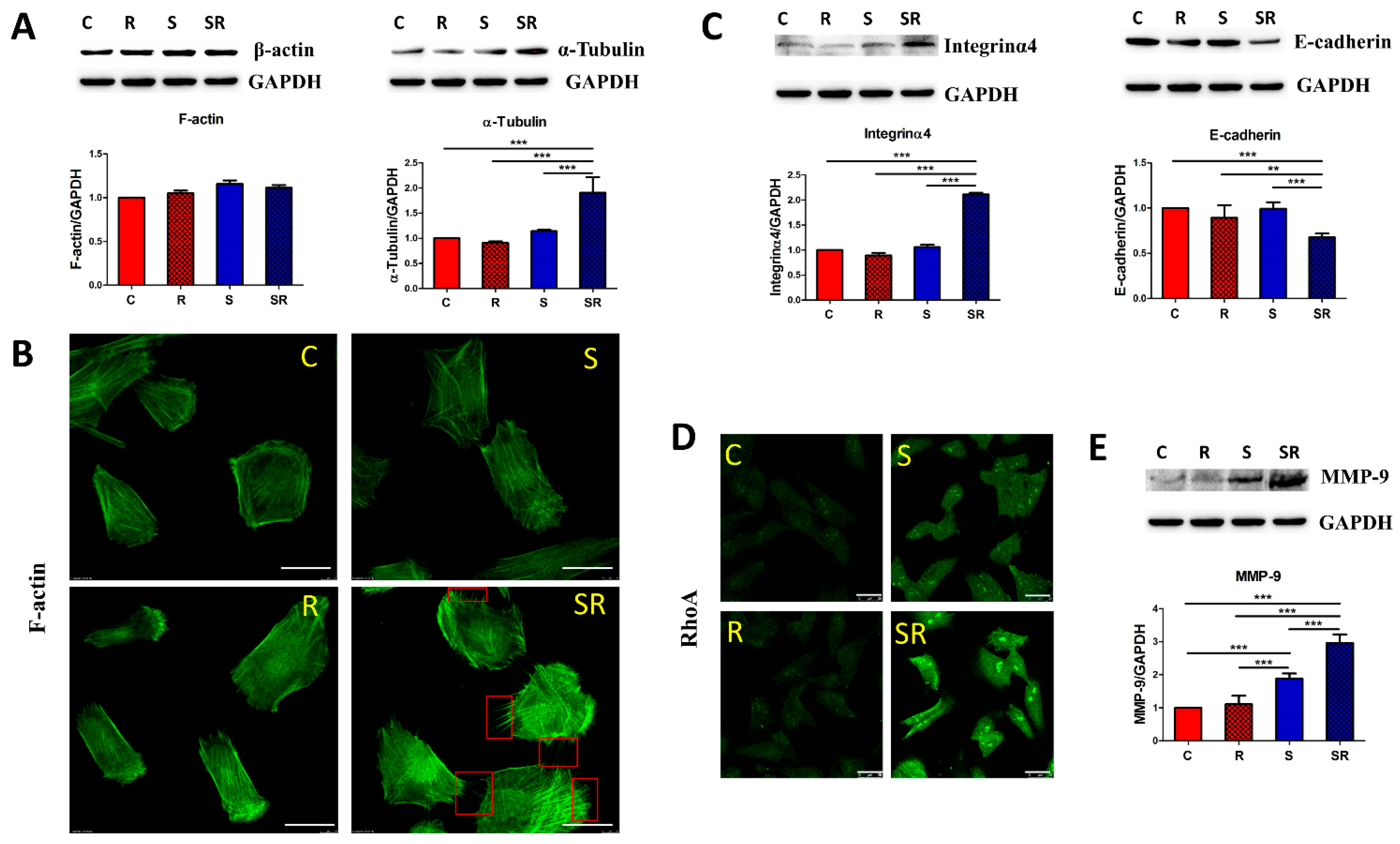
2.2. Starved and Irradiated U251 Cells Display Elevated Protein Expression of Metastatic Factors
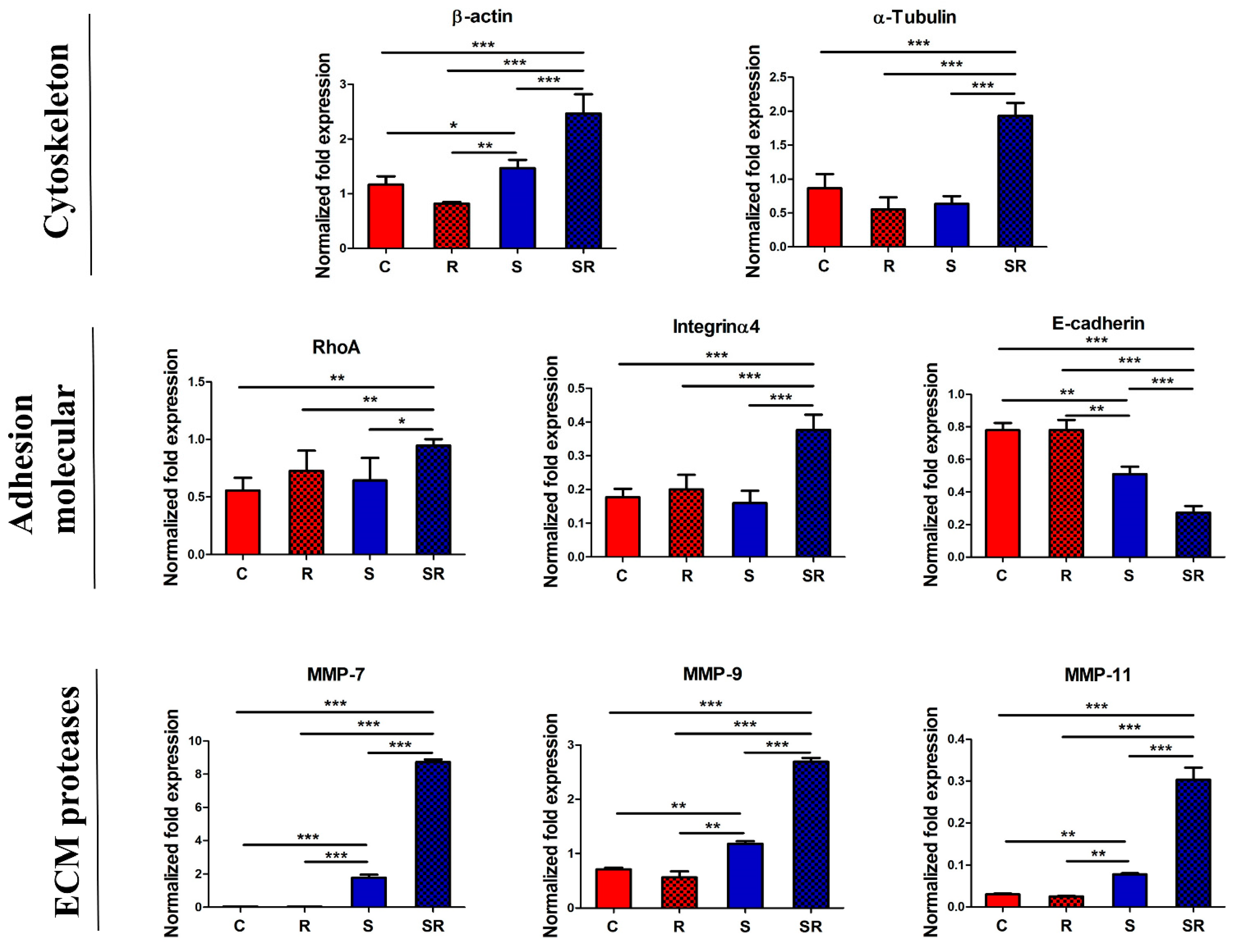
2.3. Expression of Metastatic Biomarkers Confirms the Metastatic-Inducing Ability of Starvation-Radiation Combination Treatment in U251 Cells
2.4. Starvation-Radiation Treatment Enhances U251 Metastasis by Regulating the Transcription Factor SP1
3. Discussion
4. Experimental Section
4.1. Cell Line, Cell Culture, and Treatments
4.2. High-Throughput Transcriptome Sequencing and Bioinformatics Analysis
4.3. Quantitative Real-time Reverse Transcription-PCR (Q-RT-PCR)
4.4. Western Blot Analysis
4.5. Immunofluorescence
4.6. Flow Cytometric Analysis
4.7. Metastasis Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hasegawa, M.; Asakawa, I.; Tamamoto, T.; Katayama, E.; Inoue, K.; Okada, H.; Kato, S.; Murakami, T. High-dose carbon ion beams and X-rays induce different gene expression profiles in human radioresistant glioma cells with mutant type p53 in vivo. Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, S90. [Google Scholar] [CrossRef]
- Wild-Bode, C.; Weller, M.; Rimner, A.; Dichgans, J.; Wick, W. Sublethal irradiation promotes migration and invasiveness of glioma cells: Implications for radiotherapy of human glioblastoma. Cancer Res. 2001, 61, 2744–2750. [Google Scholar] [PubMed]
- Camphausen, K.; Moses, M.A.; Beecken, W.D.; Khan, M.K.; Folkman, J.; O’Reilly, M.S. Radiation therapy to a primary tumor accelerates metastatic growth in mice. Cancer Res. 2001, 61, 2207–2211. [Google Scholar] [PubMed]
- Rolli, C.G.; Seufferlein, T.; Kemkemer, R.; Spatz, J.P. Impact of tumor cell cytoskeleton organization on invasiveness and migration: A microchannel-based approach. PLoS ONE 2010, 5, e8726. [Google Scholar] [CrossRef]
- Koukoulis, G.K.; Patriarca, C.; Gould, V.E. Adhesion molecules and tumor metastasis. Hum. Pathol. 1998, 29, 889–892. [Google Scholar] [CrossRef]
- Tang, D.G.; Honn, K.V. Adhesion molecules and tumor-metastasis—An update. Invasion Metastasis 1994, 14, 109–122. [Google Scholar] [PubMed]
- Albelda, S.M. Biology of disease—Role of integrins and other cell-adhesion molecules in tumor progression and metastasis. Lab. Investig. 1993, 68, 4–17. [Google Scholar] [PubMed]
- Beavon, I.R.G. The E-cadherin-catenin complex in tumour metastasis: Structure, function and regulation. Eur. J. Cancer 2000, 36, 1607–1620. [Google Scholar] [CrossRef]
- Stetlerstevenson, W.G.; Aznavoorian, S.; Liotta, L.A. Tumor-cell interactions with the extracellular-matrix during invasion and metastasis. Annu. Rev. Cell Biol. 1993, 9, 541–573. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Arii, S.; Mori, A.; Furutani, M.; Chiba, T.; Imamura, M. Enhancement of angiogenesis, tumor growth, and metastasis by transfection of vascular endothelial growth factor into lovo human colon cancer cell line. Clin. Cancer Res. 2000, 6, 622–630. [Google Scholar] [PubMed]
- Zheng, Q.; Liu, Y.; Zhou, H.J.; Du, Y.T.; Zhang, B.P.; Zhang, J.; Miao, G.Y.; Liu, B.; Zhang, H. X-ray radiation promotes the metastatic potential of tongue squamous cell carcinoma cells via modulation of biomechanical and cytoskeletal properties. Hum. Exp. Toxicol. 2015, 34, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Cheng, H.; Yuan, Y.; Wu, S. Regulation of ionizing radiation-induced adhesion of breast cancer cells to fibronectin by α5β1 integrin. Radiat. Res. 2014, 181, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Hamalukic, M.; Huelsenbeck, J.; Schad, A.; Wirtz, S.; Kaina, B.; Fritz, G. Rac1-regulated endothelial radiation response stimulates extravasation and metastasis that can be blocked by HMG-CoA reductase inhibitors. PLoS ONE 2011, 6, e26413. [Google Scholar] [CrossRef] [PubMed]
- Mauceri, H.J.; Hanna, N.N.; Wayne, J.D.; Hallahan, D.E.; Hellman, S.; Weichselbaum, R.R. Tumor necrosis factor alpha (TNF-α) gene therapy targeted by ionizing radiation selectively damages tumor vasculature. Cancer Res. 1996, 56, 4311–4314. [Google Scholar] [PubMed]
- Langley, R.E.; Bump, E.A.; Quartuccio, S.G.; Medeiros, D.; Braunhut, S.J. Radiation-induced apoptosis in microvascular endothelial cells. Br. J. Cancer 1997, 75, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Pohjanpelto, P.; Virtanen, I.; Holtta, E. Polyamine starvation causes disappearance of actin filaments and microtubules in polyamine-auxotrophic cho cells. Nature 1981, 293, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Kusakai, G.; Kishimoto, A.; Minegichi, Y.; Ogura, T.; Esumi, H. Induction of cell–cell detachment during glucose starvation through F-actin conversion by snark, the fourth member of the AMP-activated protein kinase catalytic subunit family. Biochem. Biophys. Res. Commun. 2003, 311, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Jean, C.; Gravelle, P.; Fournie, J.J.; Laurent, G. Influence of stress on extracellular matrix and integrin biology. Oncogene 2011, 30, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- McTyre, E.; Scott, J.; Chinnaiyan, P. Whole brain radiotherapy for brain metastasis. Surg. Neurol. Int. 2013, 4, S236–S244. [Google Scholar] [PubMed]
- Zetter, B.R. Adhesion molecules in tumor metastasis. Semin. Cancer Biol. 1993, 4, 219–229. [Google Scholar] [PubMed]
- Canel, M.; Serrels, A.; Frame, M.C.; Brunton, V.G. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J. Cell Sci. 2013, 126, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.B.; Hall, A. Rho GTPases in transformation and metastasis. Adv. Cancer Res. 2002, 84, 57–80. [Google Scholar] [PubMed]
- Cao, X.X.; Xu, J.D.; Xu, J.W.; Liu, X.L.; Cheng, Y.Y.; Li, Q.Q.; Xu, Z.D.; Liu, X.P. RACK1 promotes breast carcinoma migration/metastasis via activation of the RHoA/RHo kinase pathway. Breast Cancer Res. Treat. 2011, 126, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, H.; Ke, H.; Ni, S. MiR-129 regulates MMP9 to control metastasis of non-small cell lung cancer. Tumour Biol. 2015, 36, 5785–5790. [Google Scholar] [CrossRef] [PubMed]
- Gvozdenovic, A.; Arlt, M.J.E.; Campanile, C.; Brennecke, P.; Husmann, K.; Li, Y.F.; Born, W.; Muff, R.; Fuchs, B. CD44 enhances tumor formation and lung metastasis in experimental osteosarcoma and is an additional predictor for poor patient outcome. J. Bone Miner. Res. 2013, 28, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Klingbeil, P.; Marhaba, R.; Jung, T.; Kirmse, R.; Ludwig, T.; Zoller, M. CD44 variant isoforms promote metastasis formation by a tumor cell-matrix cross-talk that supports adhesion and apoptosis resistance. Mol. Cancer Res. 2009, 7, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Birchmeier, C.; Birchmeier, W.; Gherardi, E.; Vande Woude, G.F. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 2003, 4, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guan, X.; Zhang, J.; Jia, Z.; Wei, D.; Li, Q.; Yao, J.; Xie, K. Targeted inhibition of Sp1-mediated transcription for antiangiogenic therapy of metastatic human gastric cancer in orthotopic nude mouse models. Int. J. Oncol. 2008, 33, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Sleiman, S.F.; Langley, B.C.; Basso, M.; Berlin, J.; Xia, L.; Payappilly, J.B.; Kharel, M.K.; Guo, H.; Marsh, J.L.; Thompson, L.M.; et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J. Neurosci. 2011, 31, 6858–6870. [Google Scholar] [CrossRef] [PubMed]
- Khuntia, D.; Brown, P.; Li, J.; Mehta, M.P. Whole-brain radiotherapy in the management of brain metastasis. J. Clin. Oncol. 2006, 24, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, M. Encounter of cancer cells with bone. Radiotherapy for bone metastasis of cancer. Clin. Calcium 2011, 21, 455–464. [Google Scholar] [PubMed]
- Yoshioka, S.; Hojou, S.; Toyoda, Y.; Ogino, T.; Shimizu, K.; Yoshioka, A.; Fujie, Y.; Fukunaga, H.; Ota, H.; Endo, W.; et al. Three cases of breast cancer with skin metastasis after mastectomy treated by radiotherapy. Jpn. J. Cancer Chemother. 2010, 37, 2766–2768. [Google Scholar]
- Saito, T.; Morohashi, H.; Hasebe, T.; Sakamoto, Y.; Koyama, M.; Murata, A.; Hakamada, K. A review of stereotactic radiotherapy (SRT) for lung metastasis of colon cancer. Jpn. J. Cancer Chemother. 2014, 41, 1462–1464. [Google Scholar]
- Ward, J.F.; Evans, J.W.; Limoli, C.L.; Calabro-Jones, P.M. Radiation and hydrogen peroxide induced free radical damage to DNA. Br. J. Cancer Suppl. 1987, 8, 105–112. [Google Scholar] [PubMed]
- Nagane, M.; Yasui, H.; Yamamori, T.; Zhao, S.; Kuge, Y.; Tamaki, N.; Kameya, H.; Nakamura, H.; Fujii, H.; Inanami, O. Radiation-induced nitric oxide mitigates tumor hypoxia and radioresistance in a murine SCCVII tumor model. Biochem. Biophys. Res. Commun. 2013, 437, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.W.; Son, J.Y.; Raghavendran, H.R.; Chung, W.K.; Kim, H.G.; Park, H.J.; Jang, S.S.; Son, C.G. High-dose ionizing radiation-induced hematotoxicity and metastasis in mice model. Clin. Exp. Metastasis 2011, 28, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Sheng, S.; Li, R.; Sun, X.; Liu, J.; Huang, G. Lactate promotes resistance to glucose starvation via upregulation of Bcl-2 mediated by mTOR activation. Oncol. Rep. 2015, 33, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zou, Y.; Mao, D.; Sun, D.; Gao, G.; Shi, J.; Liu, X.; Zhu, C.; Yang, M.; Ye, W.; et al. The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J. Cell Biol. 2014, 206, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Haase, M.; Geyer, P.; Appold, S.; Schuh, D.; Kasper, M.; Muller, M. Down-regulation of Sp1 DNA binding activity in the process of radiation-induced pulmonary fibrosis. Int. J. Radiat. Biol. 2000, 76, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Ihara, T.; Tsujikawa, T.; Fujiyama, Y.; Bamba, T. Alterations in the DNA binding activity of transcriptional factors activator protein-1, Sp1, and hepatocyte nuclear factor-1 in rat jejunum during starvation and refeeding. J. Gastroenterol. Hepatol. 2003, 18, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Seznec, J.; Silkenstedt, B.; Naumann, U. Therapeutic effects of the Sp1 inhibitor mithramycin a in glioblastoma. J. Neurooncol. 2011, 101, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, J.Q.; Ouyang, S.G.; Wu, S.F.; Wang, J.; Xu, X.J.; Zhu, Y.P.; He, F.C. An integrated strategy for functional analysis in large-scale proteomic research by gene ontology. Prog. Biochem. Biophys. 2005, 32, 1026–1029. [Google Scholar]
- Prestridge, D.S. Predicting Pol II promoter sequences using transcription factor binding sites. J. Mol. Biol. 1995, 249, 923–932. [Google Scholar] [CrossRef] [PubMed]


| Name of Gene | Primer Sequence |
|---|---|
| GAPDH (f) | 5′ ACCTGCCGCCTGGAGAAACC 3′ |
| GAPDH (r) | 3′ GACCATGAGGTCCACCACCCTG 5′ |
| α-tubulin (f) | 5′ CCAAGCTGGAGTTCTCTA 3′ |
| α-tubulin (r) | 3′ CAGAGTGCTCCAGG 5′ |
| β-actin (f) | 5′ AGAGCTACGAGCTGCCTGAC 3′ |
| β-actin (r) | 3′ AGCACTGTGTTGGCGTACAG 5′ |
| RhoA (f) | 5′ CCATCATCCTGGTTGGGAAT 3′ |
| RhoA (r) | 3′ CCATGTACCCAAAAGCGC 5′ |
| E-cadherin (f) | 5′ ACCACCTCCACAGCCACCGT 3′ |
| E-cadherin (r) | 3′ GCCCACGCCAAAGTCCTCGG 5′ |
| Integrinα4 (f) | 5′ CCACCTTGGTCCTCATGTCAT 3′ |
| Integrinα4 (r) | 3′ CATGCGCAACATTCTGATCCT 5′ |
| MMP2 (f) | 5′ CGCCGTCTCCCGTCATCAAA 3′ |
| MMP2 (r) | 3′ TGAGGGTGTCCTCAGCACG 5′ |
| MMP7 (f) | 5′ TGGGCTACGTGACCTATGACAT 3′ |
| MMP7 (r) | 3′ GCCCAGCCACCTCCACTCCTC 5′ |
| MMP9 (f) | 5′ GTATGGGACATTCCTCTGATCC 3′ |
| MMP9 (r) | 3′ CCAATGAATGAATGAATGGATG 5′ |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, T.; Wang, H.; Ma, H.; Wang, H.; Chen, B.; Deng, Y. Starvation after Cobalt-60 γ-Ray Radiation Enhances Metastasis in U251 Glioma Cells by Regulating the Transcription Factor SP1. Int. J. Mol. Sci. 2016, 17, 386. https://doi.org/10.3390/ijms17040386
Zhao T, Wang H, Ma H, Wang H, Chen B, Deng Y. Starvation after Cobalt-60 γ-Ray Radiation Enhances Metastasis in U251 Glioma Cells by Regulating the Transcription Factor SP1. International Journal of Molecular Sciences. 2016; 17(4):386. https://doi.org/10.3390/ijms17040386
Chicago/Turabian StyleZhao, Tuo, Hailong Wang, Hong Ma, Hao Wang, Bo Chen, and Yulin Deng. 2016. "Starvation after Cobalt-60 γ-Ray Radiation Enhances Metastasis in U251 Glioma Cells by Regulating the Transcription Factor SP1" International Journal of Molecular Sciences 17, no. 4: 386. https://doi.org/10.3390/ijms17040386