The Human Host Defense Ribonucleases 1, 3 and 7 Are Elevated in Patients with Sepsis after Major Surgery—A Pilot Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. Study Population
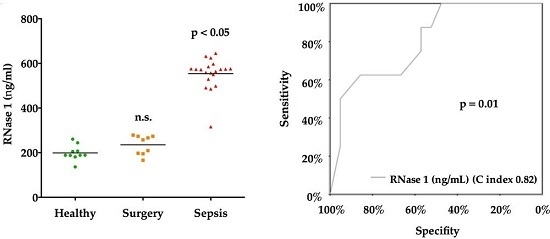
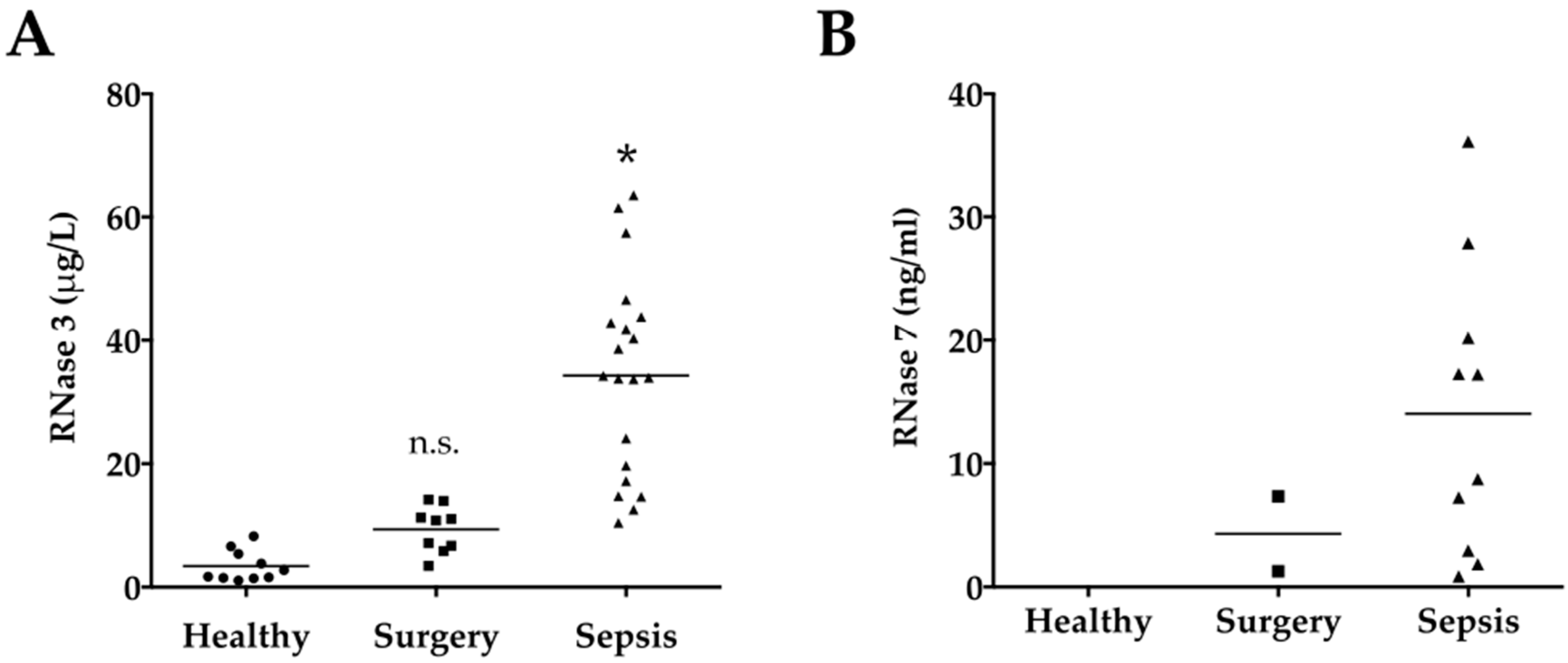
2.2. Serum Levels of RNases
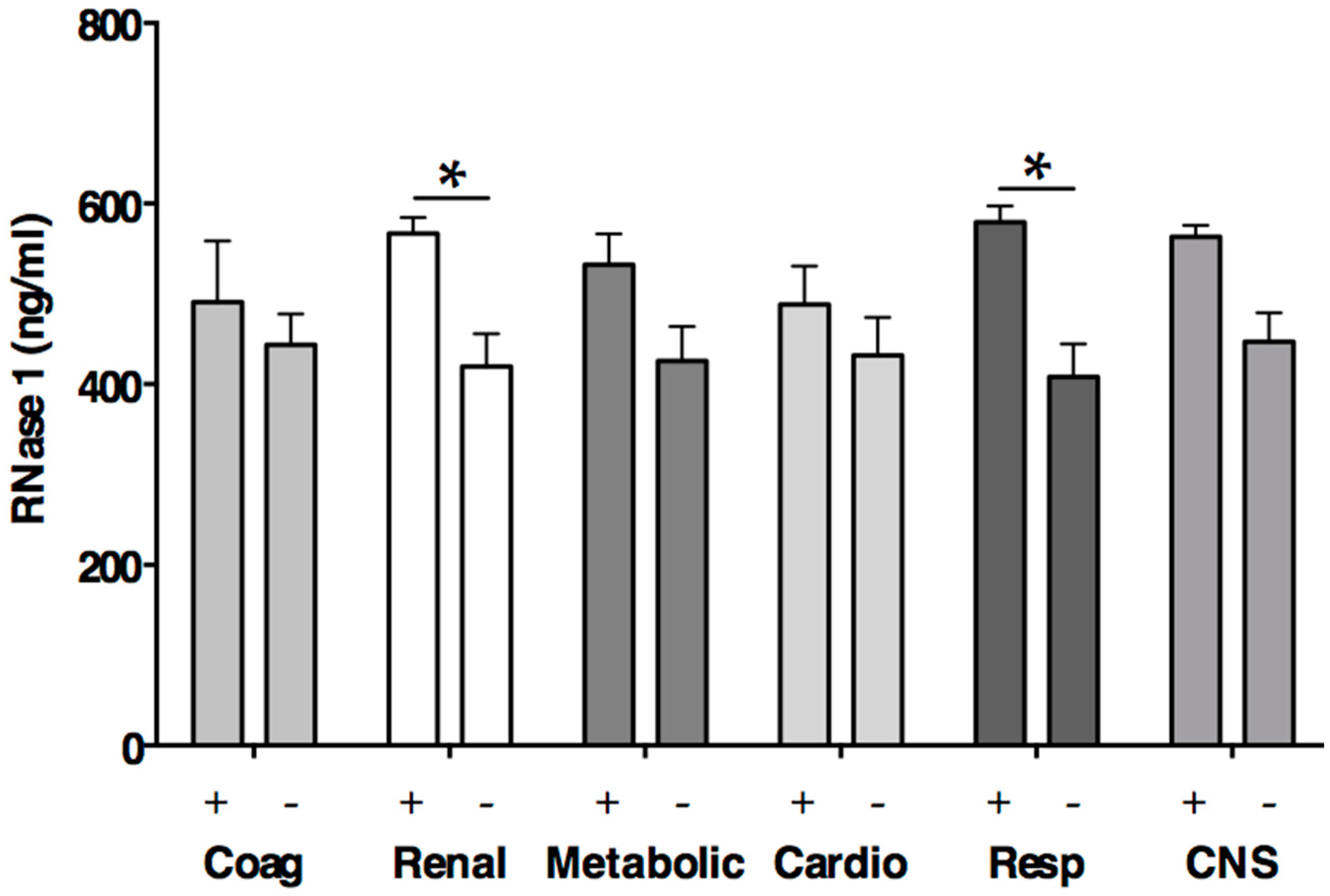
2.3. RNase 1
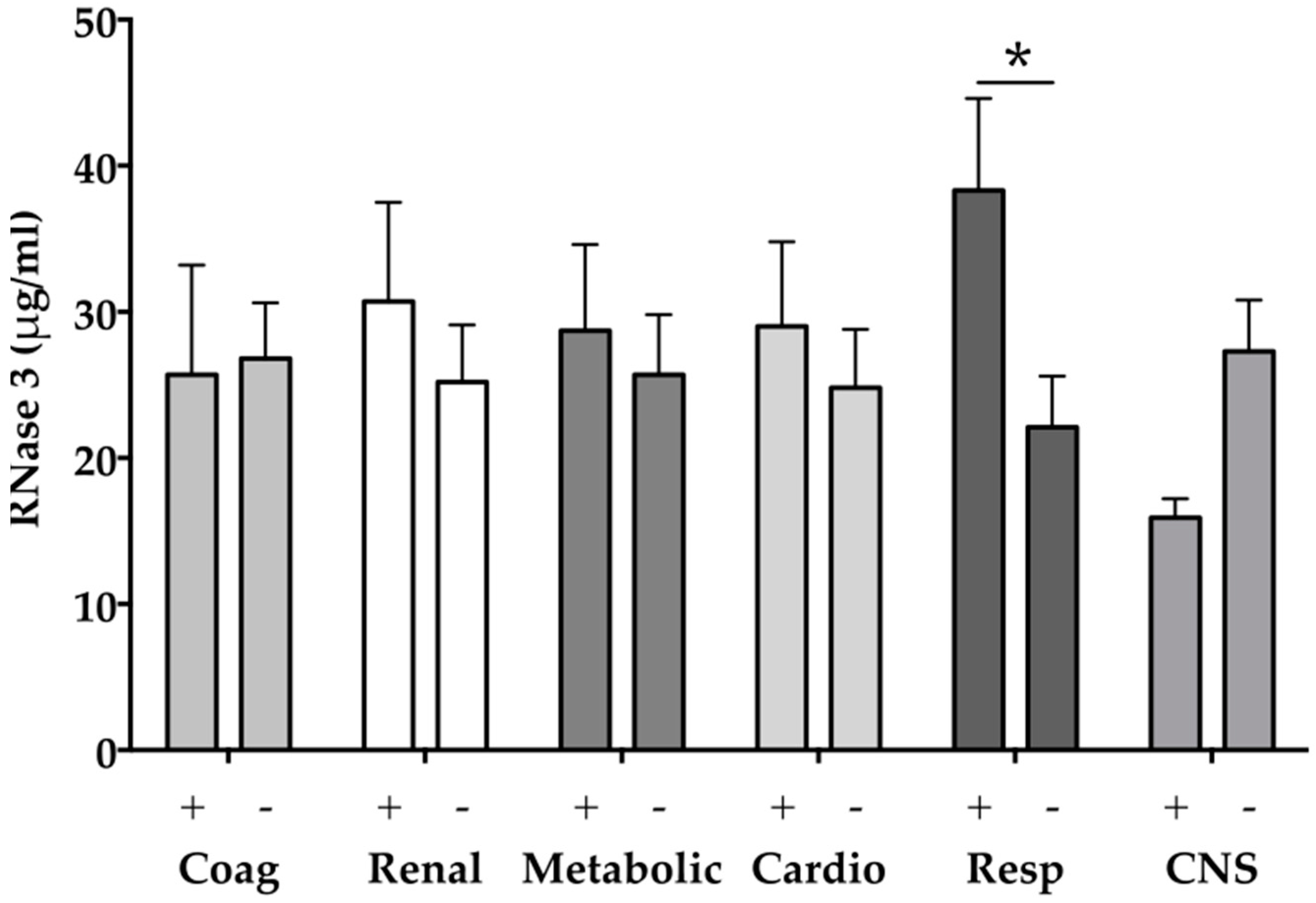
2.4. RNase 3
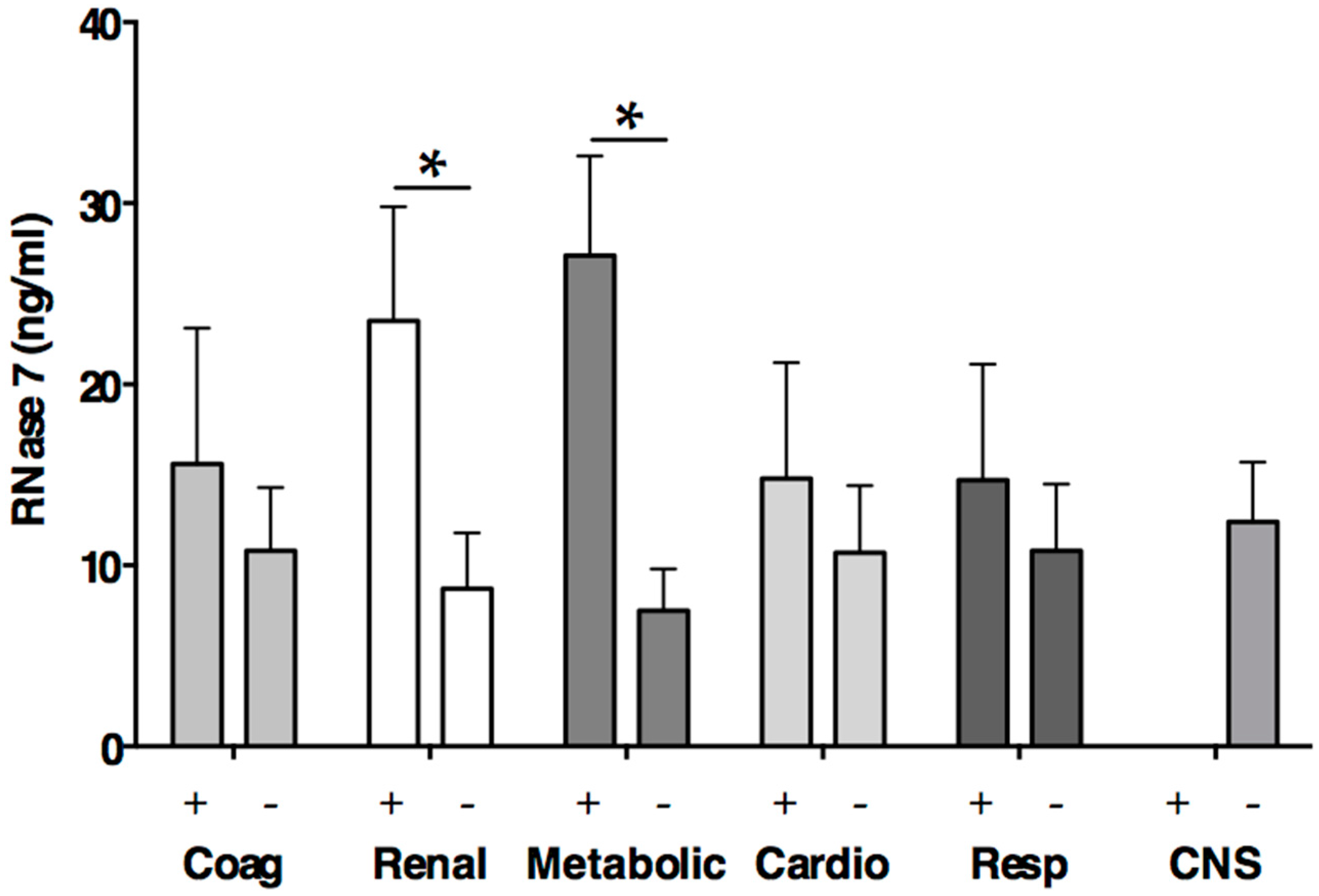
2.5. RNase 7
3. Materials and Methods
3.1. Ethical Approach
3.2. Study Population
3.3. Data Collection
3.4. Sample Collection
3.5. Ribonucelases–Enzyme-Linked Immunosorbent Assays
3.6. Statistical Analyses
Author Contributions
Conflicts of Interest
Abbreviations
| AMP | antimicrobial peptide |
| APACHE II | Acute Physiology and Chronic Health Evaluation II score |
| BMI | body-mass-index |
| CRP | C-reactive protein |
| ECP | eosinophil cationic peptide |
| EDN | eosinophil-derived cationic protein |
| ELISA | enzyme linked immunoassay |
| ICU | intensive care unit |
| IL-1 β | interleukin 1β |
| LOS | length of stay |
| MV | mechanical ventilation |
| PCT | procalcitonin |
| RNase | ribonucleases |
| SCCM | Society of Critical Care Medicine |
| SOFA | Sequential Organ Failure Assessment score |
| TNF-α | tumor necrosis factor alpha |
References
- Vincent, J.-L.; Rello, J.; Marshall, J.; Silva, E.; Anzueto, A.; Martin, C.D.; Moreno, R.; Lipman, J.; Gomersall, C.; Sakr, Y.; et al. EPIC II Group of investigators international study of the prevalence and outcomes of infection in intensive care units. JAMA J. Am. Med. Assoc. 2009, 302, 2323–2329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rittirsch, D.; Flierl, M.A.; Ward, P.A. Harmful molecular mechanisms in sepsis. Nat. Rev. Immunol. 2008, 8, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. The immunopathogenesis of sepsis. Nature 2002, 420, 885–891. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Ganz, T. Antimicrobial peptides in mammalian and insect host defence. J. Surg. Res. 1999, 11, 23–27. [Google Scholar] [CrossRef]
- Martin, L.; van Meegern, A.; Doemming, S.; Schuerholz, T. Antimicrobial peptides in human sepsis. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Haigh, B.J.; Griffin, F.J.; Wheeler, T.T. The mammalian secreted RNases: Mechanisms of action in host defence. Innate Immun. 2013, 19, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Confalone, E.; Breukelman, H.J.; Sasso, M.P.; Jekel, P.A.; Hodge, E.; Furia, A.; Beintema, J.J. Ruminant brain ribonucleases: Expression and evolution. Biochim. Biophys. Acta 2001, 1547, 95–103. [Google Scholar] [CrossRef]
- Sorrentino, S.; Glitz, D.G.; Hamann, K.J.; Loegering, D.A.; Checkel, J.L.; Gleich, G.J. Eosinophil-derived neurotoxin and human liver ribonuclease. Identity of structure and linkage of neurotoxicity to nuclease activity. J. Biol. Chem. 1992, 267, 14859–14865. [Google Scholar] [PubMed]
- Spencer, J.D.; Schwaderer, A.L.; DiRosario, J.D.; McHugh, K.M.; McGillivary, G.; Justice, S.S.; Carpenter, A.R.; Baker, P.B.; Harder, J.; Hains, D.S. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 2011, 80, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Cormier, S.A.; Larson, K.A.; Yuan, S.; Mitchell, T.L.; Lindenberger, K.; Carrigan, P.; Lee, N.A.; Lee, J.J. Mouse eosinophil-associated ribonucleases: A unique subfamily expressed during hematopoiesis. Mamm. Genome 2001, 12, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, S.J.; Loegering, D.A.; Venge, P.; Olsson, I.; Harley, J.B.; Fauci, A.S.; Gleich, G.J. Distinctive cationic proteins of the human eosinophil granule: Major basic protein, eosinophil cationic protein, and eosinophil-derived neurotoxin. J. Immunol. (Baltimore Md. 1950) 1983, 131, 2977–2982. [Google Scholar]
- Durack, D.T.; Ackerman, S.J.; Loegering, D.A.; Gleich, G.J. Purification of human eosinophil-derived neurotoxin. Proc. Natl. Acad. Sci. USA 1981, 78, 5165–5169. [Google Scholar] [CrossRef] [PubMed]
- Egesten, A.; Dyer, K.D.; Batten, D.; Domachowske, J.B.; Rosenberg, H.F. Ribonucleases and host defense: Identification, localization and gene expression in adherent monocytes in vitro. Biochim. Biophys. Acta 1997, 1358, 255–260. [Google Scholar] [CrossRef]
- Cormier, S.A.; Yuan, S.; Crosby, J.R. TH2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 2002, 27, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Beintema, J.J.; Kleineidam, R.G. The ribonuclease a superfamily: General discussion. Cell. Mol. Life Sci. CMLS 1998, 54, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, S. The eight human “canonical” ribonucleases: Molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010, 584, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Landré, J.B.P.; Hewett, P.W.; Olivot, J.-M.; Friedl, P.; Ko, Y.; Sachinidis, A.; Moenner, M. Human endothelial cells selectively express large amounts of pancreatic-type ribonuclease (RNase 1). J. Cell. Biochem. 2002, 86, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, S.; Naddeo, M.; Russo, A.; D’Alessio, G. Degradation of double-stranded RNA by human pancreatic ribonuclease: Crucial role of noncatalytic basic amino acid residues. Biochemistry 2003, 42, 10182–10190. [Google Scholar] [CrossRef] [PubMed]
- De, Y.; Chen, Q.; Rosenberg, H.F.; Rybak, S.M.; Newton, D.L.; Wang, Z.Y.; Fu, Q.; Tchernev, V.T.; Wang, M.; Schweitzer, B.; et al. Ribonuclease a superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. J. Immunol. (Baltimore Md. 1950) 2004, 173, 6134–6142. [Google Scholar]
- Shamri, R.; Xenakis, J.J.; Spencer, L.A. Eosinophils in innate immunity: An evolving story. Cell Tissue Res. 2011, 343, 57–83. [Google Scholar] [CrossRef] [PubMed]
- Niimi, A.; Amitani, R.; Suzuki, K.; Tanaka, E.; Murayama, T.; Kuze, F. Serum eosinophil cationic protein as a marker of eosinophilic inflammation in asthma. Clin. Exp. Allergy 1998, 28, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Blom, K.; Rubin, J.; Halfvarson, J.; Törkvist, L.; Rönnblom, A.; Sangfelt, P.; Lördal, M.; Jönsson, U.-B.; Sjöqvist, U.; Håkansson, L.D.; et al. Eosinophil associated genes in the inflammatory bowel disease 4 region: Correlation to inflammatory bowel disease revealed. World J. Gastroenterol. 2012, 18, 6409–6419. [Google Scholar] [CrossRef] [PubMed]
- Carreras, E.; Boix, E.; Rosenberg, H.F.; Cuchillo, C.M.; Nogués, M.V. Both aromatic and cationic residues contribute to the membrane-lytic and bactericidal activity of eosinophil cationic protein. Biochemistry 2003, 42, 6636–6644. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Navarro, S.; Moussaoui, M.; Nogués, M.V.; Boix, E. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 2008, 47, 3544–3555. [Google Scholar] [CrossRef] [PubMed]
- Navarro, S.; Aleu, J.; Jiménez, M.; Boix, E.; Cuchillo, C.M.; Nogues, M.V. The cytotoxicity of eosinophil cationic protein/ribonuclease 3 on eukaryotic cell lines takes place through its aggregation on the cell membrane. CMLS Cell. Mol. Life Sci. 2008, 65, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.C.; Lo, C.W.; Fan, T.; Chang, M. TNF-α mediates eosinophil cationic protein-induced apoptosis in BEAS-2B cells. BMC Cell Biol. 2010, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Schröder, J.M. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002, 277, 46779–46784. [Google Scholar] [CrossRef] [PubMed]
- Abtin, A.; Eckhart, L.; Mildner, M.; Ghannadan, M. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. J. Investig. Dermatol. 2009, 129, 2193–2201. [Google Scholar] [CrossRef] [PubMed]
- Laudien, M.; Dressel, S.; Harder, J.; Gläser, R. Differential expression pattern of antimicrobial peptides in nasal mucosa and secretion. Rhinology 2011, 49, 107–111. [Google Scholar] [PubMed]
- Spencer, J.D.; Schwaderer, A.L.; Wang, H.; Bartz, J.; Kline, J.; Eichler, T.; DeSouza, K.R.; Sims-Lucas, S.; Baker, P.; Hains, D.S. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. 2013, 83, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Wiese, A.; Brandenburg, K.; Carroll, S.F.; Rietschel, E.T.; Seydel, U. Mechanisms of action of bactericidal/permeability-increasing protein BPI on reconstituted outer membranes of gram-negative bacteria. Biochemistry 1997, 36, 10311–10319. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-M.; Wu, S.-J.; Chang, T.-W.; Wang, C.-F.; Suen, C.-S.; Hwang, M.-J.; Chang, M.D.-T.; Chen, Y.-T.; Liao, Y.-D. Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein. J. Biol. Chem. 2010, 285, 8985–8994. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Weickmann, J.L.; Olson, E.M.; Glitz, D.G. Immunological assay of pancreatic ribonuclease in serum as an indicator of pancreatic cancer. Cancer Res. 1984, 44, 1682–1687. [Google Scholar]
- Kannemeier, C.; Shibamiya, A.; Nakazawa, F.; Trusheim, H.; Ruppert, C.; Markart, P.; Song, Y.; Tzima, E.; Kennerknecht, E.; Niepmann, M.; et al. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc. Natl. Acad. Sci. USA 2007, 104, 6388–6393. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Gerriets, T.; Wessels, C.; Walberer, M.; Kostin, S.; Stolz, E.; Zheleva, K.; Hocke, A.; Hippenstiel, S.; Preissner, K.T. Extracellular RNA mediates endothelial-cell permeability via vascular endothelial growth factor. Blood 2007, 110, 2457–2465. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Grantzow, T.; Pagel, J.I.; Tschernatsch, M.; Sperandio, M.; Preissner, K.T.; Deindl, E. Extracellular RNA promotes leukocyte recruitment in the vascular system by mobilising proinflammatory cytokines. Thromb. Haemost. 2012, 108, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Gansler, J.; Preissner, K.T.; Fischer, S. Influence of proinflammatory stimuli on the expression of vascular ribonuclease 1 in endothelial cells. FASEB J. 2014, 28, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving sepsis campaign guidelines committee including the pediatric subgroup surviving sepsis campaign: International guidelines for management of severe sepsis and septic shock. Intensive Care Med. 2013, 39, 165–228. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, R.L.; Karpetsky, T.P.; Neuwelt, E.A.; Levy, C.C. Levels of serum ribonuclease as an indicator of renal insufficiency in patients with leukemia. Cancer Res. 1977, 37, 2015–2022. [Google Scholar] [PubMed]
- Yousefi, S.; Gold, J.A.; Andina, N.; Lee, J.J.; Kelly, A.M. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 2008, 14, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Abidi, K.; Khoudri, I.; Belayachi, J.; Madani, N.; Zekraoui, A.; Zeggwagh, A.A.; Abouqal, R. Eosinopenia is a reliable marker of sepsis on admission to medical intensive care units. Crit. Care 2008, 12, R59. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, U.-B.; Blom, K.; Stålenheim, G.; Håkansson, L.D.; Venge, P. The production of the eosinophil proteins ECP and EPX/EDN are regulated in a reciprocal manner. APMIS 2014, 122, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Amin, K.; Lúdvíksdóttir, D.; Janson, C.; Nettelbladt, O.; Bjornsson, E.; Roomans, G.M.; Boman, G.; Sevéus, L.; Venge, P. Inflammation and structural changes in the airways of patients with atopic and nonatopic asthma. BHR Group. Am. J. Respir. Crit. Care Med. 2000, 162, 2295–2301. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Kitahara, H.; Kanemitsu, T. Eosinophil cationic protein in the sera of patients with Mycoplasma pneumonia. Pediatr. Infect. Dis. J. 1994, 13, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Bagshaw, S.M.; Lapinsky, S.; Dial, S.; Arabi, Y.; Dodek, P.; Wood, G.; Ellis, P.; Guzman, J.; Marshall, J.; Parrillo, J.E.; et al. Acute kidney injury in septic shock: Clinical outcomes and impact of duration of hypotension prior to initiation of antimicrobial therapy. Intensive Care Med. 2008, 35, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Casserly, B.; Phillips, G.S.; Schorr, C.; Dellinger, R.P.; Townsend, S.R.; Osborn, T.M.; Reinhart, K.; Selvakumar, N.; Levy, M.M. Lactate measurements in sepsis-induced tissue hypoperfusion: Results from the Surviving Sepsis Campaign database. Crit. Care Med. 2015, 43, 567–573. [Google Scholar] [CrossRef] [PubMed]

| Healthy (n = 10) | Surgery (n = 9) | Sepsis (n = 20) | p-Value | |
|---|---|---|---|---|
| Age (years) (IQR) | 55 (49–74) | 70 (55–75) | 64 (55–75) | 0.63 |
| Male sex (%) | 7 (70.0) | 4 (44.4) | 12 (60.0) | 0.52 |
| BMI (kg/m2) (IQR) | - | 24.3 (22.1–35.1) | 27.3 (30.9–22.6) | 0.72 |
| Diabetes mellitus (%) | - | 3 (33.3) | 4 (20.0) | 0.5 |
| Creatinine (mg/dL) (IQR) | - | 0.9 (0.6–1.0) | 0.8 (0.7–1.3) | 0.71 |
| Hemoglobin (g/dL) (IQR) | - | 11.3 (10.1–12.0) | 9.3 (8.4–9.6) | <0.0001 |
| Platelets (109 cells/nL) (IQR) | - | 255.0 (168.0–296.0) | 193.5 (173.0–257.8) | 0.32 |
| White cells (109 cells/nL) (IQR) | - | 10.5 (8.8–12.5) | 17.5 (14.3–21.1) | 0.02 |
| Neutrophil (109 cells/nL) (IQR) | - | 85.5 (84.9–87.9) | 86.0 (76.5–93.0) | 0.89 |
| Eosinophil (109 cells/nL) (IQR) | - | 0.2 (0.1–0.2) | 0.0 (0.0–1.5) | 0.69 |
| Monocyte (109 cells/nL) (IQR) | - | 4.9 (2.5–6.4) | 4.3 (3.5–6.0) | 0.91 |
| Lymphocyte (109 cells/nL) (IQR) | - | 9.3 (4.1–11.7) | 5.0 (2.9–10.5) | 0.26 |
| Albumin (g/L) (IQR) | - | 31.0 (26.5–38.5) | 22.0 (19.0–26.0) | <0.0001 |
| PCT (ng/mL) (IQR) | - | 0.1 (0.0– 0.2) | 3.1 (0.6–21.8) | <0.0001 |
| CRP (mg/dL) (IQR) | - | 6.6 (2.8– 25.0) | 170.5 (113.0–230.0) | <0.0001 |
| Lactate (mmol/L) (IQR) | - | 0.9 (0.8–1.7) | 1.5 (1.2–2.5) | 0.05 |
| Fluid administration within first 24 h (L) (IQR) | - | 4.9 (3.0–5.0) | 5.2 (3.5–7.0) | 0.13 |
| Urine output within first 24 h (L) (IQR) | - | 1.9 (1.3–2.3) | 3.4 (2.6–4.3) | <0.0001 |
| SOFA (points) (IQR) | - | 5.0 (0.5–6.0) | 6.0 (3.3–8.0) | 0.06 |
| APACHE II (points) (IQR) | - | 9.0 (4.0–10.5) | 11.5 (8.5–16.8) | 0.03 |
| Vasopressors (h) (IQR) | - | 2.5 (0.0–31.0) | 30.0 (2.0–68.8) | 0.11 |
| MV (h) (IQR) | - | 3.0 (0.3–7.0) | 3.0 (0.0–45.5) | 0.55 |
| LOS ICU (days) (IQR) | - | 1.0 (1.0–3.0) | 7.0 (5.0–12.0) | <0.0001 |
| 28-day mortality (%) | - | 0 (0.0) | 0 (0.0) | - |
| Healthy (n = 10) | Surgery (n = 9) | Sepsis (n = 20) | p-Value | |
|---|---|---|---|---|
| RNase 1 (ng/mL) (IQR) | 188.7 (188.0–216.2) | 257.5 (196.2–273.3) | 572.6 (534.5–582.9) | <0.0001 |
| RNase 3 (μg/mL) (IQR) | 2.2 (1.5–5.7) | 10.8 (6.3–12.6) | 34.1 (17.8–43.5) | <0.0001 |
| RNase 7 (ng/mL) (IQR) | n.d. | 4.3 (1.0–7.3) # | 13.0 (2.7–22.1) ## | 0.28 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, L.; Koczera, P.; Simons, N.; Zechendorf, E.; Hoeger, J.; Marx, G.; Schuerholz, T. The Human Host Defense Ribonucleases 1, 3 and 7 Are Elevated in Patients with Sepsis after Major Surgery—A Pilot Study. Int. J. Mol. Sci. 2016, 17, 294. https://doi.org/10.3390/ijms17030294
Martin L, Koczera P, Simons N, Zechendorf E, Hoeger J, Marx G, Schuerholz T. The Human Host Defense Ribonucleases 1, 3 and 7 Are Elevated in Patients with Sepsis after Major Surgery—A Pilot Study. International Journal of Molecular Sciences. 2016; 17(3):294. https://doi.org/10.3390/ijms17030294
Chicago/Turabian StyleMartin, Lukas, Patrick Koczera, Nadine Simons, Elisabeth Zechendorf, Janine Hoeger, Gernot Marx, and Tobias Schuerholz. 2016. "The Human Host Defense Ribonucleases 1, 3 and 7 Are Elevated in Patients with Sepsis after Major Surgery—A Pilot Study" International Journal of Molecular Sciences 17, no. 3: 294. https://doi.org/10.3390/ijms17030294