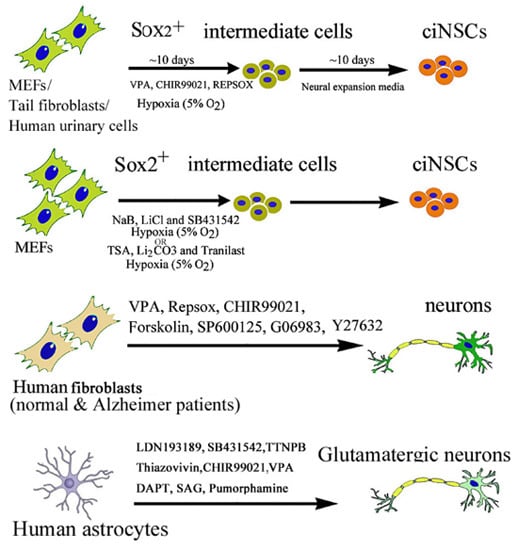
Chemically Induced Reprogramming of Somatic Cells to Pluripotent Stem Cells and Neural Cells
Abstract
:1. Introduction
2. Chemically Induced Reprogramming of Somatic Cells
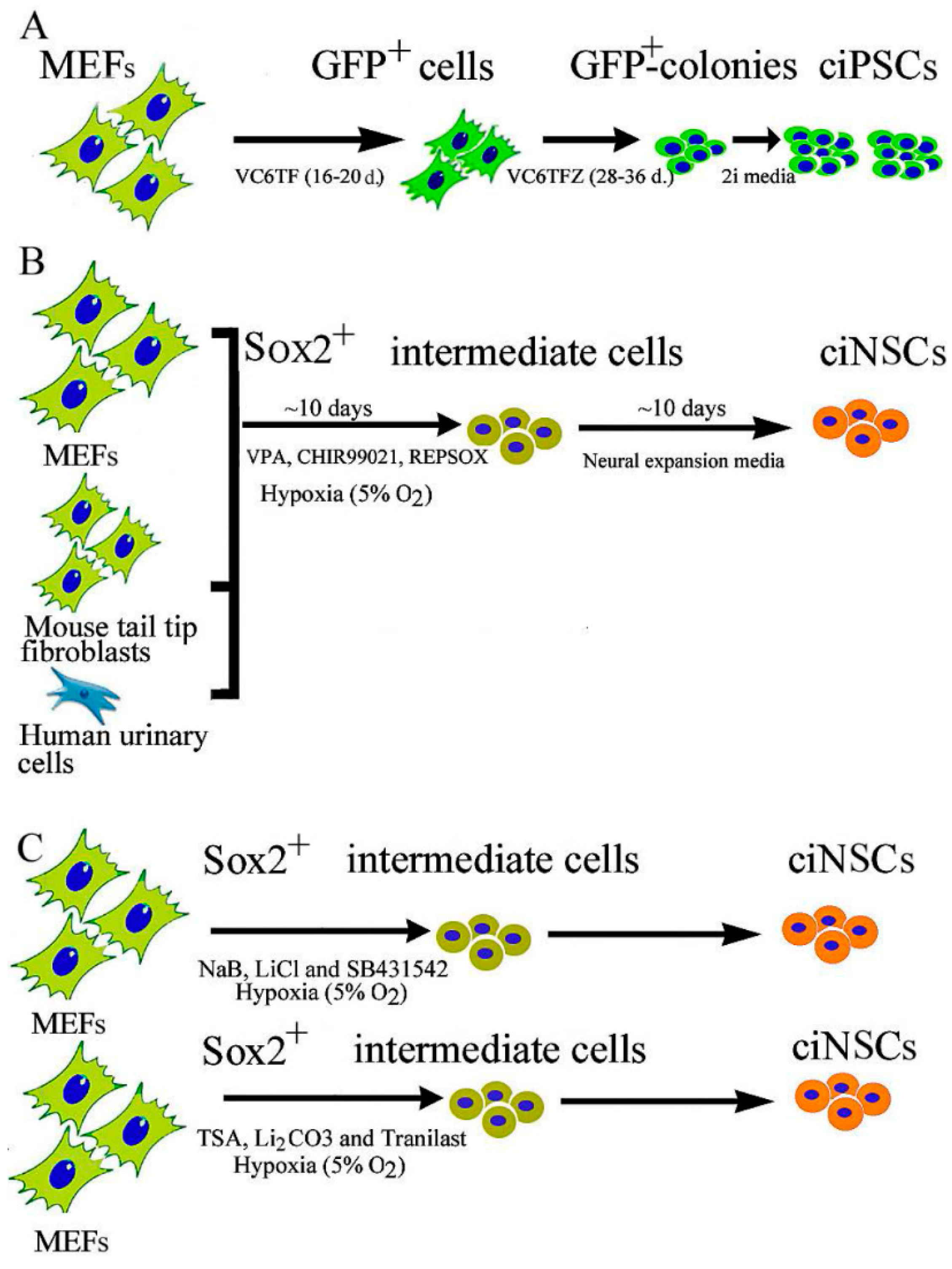
2.1. Chemical-Induced Generation of iPSCs
2.2. Chemical-Induced Generation of Neural Stem Cells
2.3. Chemical-Induced Generation of Neurons

3. Mechanisms of Actions of Small Molecules in the Context of iPSC and Neural Cell Reprogramming
3.1. Modulators of DNA Methylation
3.2. Modulators of Histone Methylation
3.3. Modulators of Histone Acetylation
3.4. Modulation of Cell Signaling
4. Conclusions
Author Contributions
Conflicts of Interest
References
- Kriegstein, A.; Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009, 32, 149–184. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Teng, E.M.; Summers, R.G., Jr.; Ming, G.L.; Gage, F.H. Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J. Neurosci. 2006, 26, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Horner, P.J.; Power, A.E.; Kempermann, G.; Kuhn, H.G.; Palmer, T.D.; Winkler, J.; Thal, L.J.; Gage, F.H. Proliferation and differentiation of progenitor cells throughout the intact adult rat spinal cord. J. Neurosci. 2000, 20, 2218–2228. [Google Scholar] [PubMed]
- Nishiyama, A.; Komitova, M.; Suzuki, R.; Zhu, X. Polydendrocytes (NG2 cells): Multifunctional cells with lineage plasticity. Nat. Rev. Neurosci. 2009, 10, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chan, A.; Wen, H.; Chung, S.H.; Deng, W.; Jiang, P. Stem and progenitor cell-derived astroglia therapies for neurological diseases. Trends Mol. Med. 2015, 21, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, V.; Jiang, P.; Chechneva, O.; Lo, U.G.; Deng, W. Differentiating human stem cells into neurons and glial cells for neural repair. Front. Biosci. 2012, 17, 65–89. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, E.Y.; Ichida, J.K.; Wainger, B.J.; Toma, J.S.; Rafuse, V.F.; Woolf, C.J.; Eggan, K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 2011, 9, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Sudhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zuchero, J.B.; Ahlenius, H.; Marro, S.; Ng, Y.H.; Vierbuchen, T.; Hawkins, J.S.; Geissler, R.; Barres, B.A.; Wernig, M. Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 2013, 31, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Najm, F.J.; Lager, A.M.; Zaremba, A.; Wyatt, K.; Caprariello, A.V.; Factor, D.C.; Karl, R.T.; Maeda, T.; Miller, R.H.; Tesar, P.J. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat. Biotechnol. 2013, 31, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wang, J.; Jia, J.; Song, N.; Xiang, C.; Xu, J.; Hou, Z.; Su, X.; Liu, B.; Jiang, T.; et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 2014, 14, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.; Osafune, K.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Chen, S.; Muhlestein, W.; Melton, D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008, 26, 1269–1275. [Google Scholar] [CrossRef] [PubMed]
- Armond, J.W.; Saha, K.; Rana, A.A.; Oates, C.J.; Jaenisch, R.; Nicodemi, M.; Mukherjee, S. A stochastic model dissects cell states in biological transition processes. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Desponts, C.; Do, J.T.; Hahm, H.S.; Scholer, H.R.; Ding, S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 2008, 3, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Do, J.T.; Desponts, C.; Hahm, H.S.; Scholer, H.R.; Ding, S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2008, 2, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Ambasudhan, R.; Yuan, X.; Li, W.; Hilcove, S.; Abujarour, R.; Lin, X.; Hahm, H.S.; Hao, E.; Hayek, A.; et al. A chemical platform for improved induction of human iPSCs. Nat. Methods 2009, 6, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Yin, X.; Yang, W.; Du, Y.; Hou, P.; Ge, J.; Liu, C.; Zhang, W.; Zhang, X.; et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011, 21, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wan, H.; Zhao, X.; Zhu, S.; Zhou, Q.; Ding, S. Brief report: Combined chemical treatment enables Oct4-induced reprogramming from mouse embryonic fibroblasts. Stem Cells 2011, 29, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 2010, 7, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K.; et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Elite and stochastic models for induced pluripotent stem cell generation. Nature 2009, 460, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Ladewig, J.; Koch, P.; Brustle, O. Leveling Waddington: The emergence of direct programming and the loss of cell fate hierarchies. Nat. Rev. Mol. Cell Biol 2013, 14, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, T.; Guan, J.; Zhang, X.; Fu, Y.; Ye, J.; Zhu, J.; Meng, G.; Ge, J.; Yang, S.; et al. A xen-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell 2015, 163, 1678–1691. [Google Scholar] [CrossRef] [PubMed]
- Gage, F.H.; Temple, S. Neural stem cells: Generating and regenerating the brain. Neuron 2013, 80, 588–601. [Google Scholar] [CrossRef] [PubMed]
- Han, D.W.; Tapia, N.; Hermann, A.; Hemmer, K.; Hoing, S.; Arauzo-Bravo, M.J.; Zaehres, H.; Wu, G.; Frank, S.; Moritz, S.; et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 2012, 10, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Ring, K.L.; Tong, L.M.; Balestra, M.E.; Javier, R.; Andrews-Zwilling, Y.; Li, G.; Walker, D.; Zhang, W.R.; Kreitzer, A.C.; Huang, Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 2012, 11, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Hu, W.; Qiu, B.; Zhao, J.; Yu, Y.; Guan, W.; Wang, M.; Yang, W.; Pei, G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014, 24, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zuo, X.; Jing, J.; Ma, Y.; Wang, J.; Liu, D.; Zhu, J.; Du, X.; Xiong, L.; Du, Y.; et al. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 2015, 17, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human es and ips cells by dual inhibition of smad signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Ladewig, J.; Mertens, J.; Kesavan, J.; Doerr, J.; Poppe, D.; Glaue, F.; Herms, S.; Wernet, P.; Kogler, G.; Muller, F.J.; et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 2012, 9, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Seamon, K.B.; Padgett, W.; Daly, J.W. Forskolin: Unique diterpene activator of adenylate cyclase in membranes and in intact cells. Proc. Natl. Acad. Sci. USA 1981, 78, 3363–3367. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Qiu, B.; Guan, W.; Wang, Q.; Wang, M.; Li, W.; Gao, L.; Shen, L.; Huang, Y.; Xie, G.; et al. Direct conversion of normal and alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 2015, 17, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.L.; Zang, T.; Zou, Y.; Chang, J.C.; Gibson, J.R.; Huber, K.M.; Zhang, C.L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lamas, N.J.; Johnson-Kerner, B.; Roybon, L.; Kim, Y.A.; Garcia-Diaz, A.; Wichterle, H.; Henderson, C.E. Neurotrophic requirements of human motor neurons defined using amplified and purified stem cell-derived cultures. PLoS ONE 2014, 9, e110324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yin, J.C.; Yeh, H.; Ma, N.X.; Lee, G.; Chen, X.A.; Wang, Y.; Lin, L.; Chen, L.; Jin, P.; et al. Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell 2015, 17, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Takai, D. The role of DNA methylation in mammalian epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Hanna, J.; Zhang, X.; Ku, M.; Wernig, M.; Schorderet, P.; Bernstein, B.E.; Jaenisch, R.; Lander, E.S.; Meissner, A. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008, 454, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Bickenbach, J.R.; Tomanek-Chalkley, A.; Wiechert, S.; Winter, M.C. Human skin keratinocytes can be reprogrammed to express neuronal genes and proteins after a single treatment with decitabine. Biores. Open Access 2013, 2, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Chinnathambi, S.; Wiechert, S.; Tomanek-Chalkley, A.; Winter, M.C.; Bickenbach, J.R. Treatment with the cancer drugs decitabine and doxorubicin induces human skin keratinocytes to express Oct4 and the OCT4 regulator mir-145. J. Dermatol. 2012, 39, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Fu, C.; Shen, C.; Shi, Y. Histone deacetylases in neural stem cells and induced pluripotent stem cells. J. Biomed. Biotechnol. 2011, 2011, 835968. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Adjaye, J. A cyclic AMP analog, 8-Br-cAMP, enhances the induction of pluripotency in human fibroblast cells. Stem Cell Rev. 2011, 7, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Gao, F.; Jin, Y. The regulatory role of histone deacetylase inhibitors in Fgf4 expression is dependent on the differentiation state of pluripotent stem cells. J. Cell. Physiol. 2011, 226, 3190–3196. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Chen, X.; Yu, D.; Li, T.; Cui, J.; Wang, G.; Hu, J.F.; Li, W. Histone deacetylase inhibitor valproic acid promotes the induction of pluripotency in mouse fibroblasts by suppressing reprogramming-induced senescence stress. Exp. Cell Res. 2015, 337, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Wattanapanitch, M.; Klincumhom, N.; Potirat, P.; Amornpisutt, R.; Lorthongpanich, C.; U-pratya, Y.; Laowtammathron, C.; Kheolamai, P.; Poungvarin, N.; Issaragrisil, S. Dual small-molecule targeting of smad signaling stimulates human induced pluripotent stem cells toward neural lineages. PLoS ONE 2014, 9, e106952. [Google Scholar] [CrossRef] [PubMed]
- Pandian, G.N.; Nakano, Y.; Sato, S.; Morinaga, H.; Bando, T.; Nagase, H.; Sugiyama, H. A synthetic small molecule for rapid induction of multiple pluripotency genes in mouse embryonic fibroblasts. Sci. Rep. 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandian, G.N.; Sato, S.; Anandhakumar, C.; Taniguchi, J.; Takashima, K.; Syed, J.; Han, L.; Saha, A.; Bando, T.; Nagase, H.; et al. Identification of a small molecule that turns on the pluripotency gene circuitry in human fibroblasts. ACS Chem. Biol. 2014, 9, 2729–2736. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.H.; Ho, J.C.; Lee, Y.K.; Ng, K.M.; Au, K.W.; Chan, Y.C.; Lau, C.P.; Tse, H.F.; Siu, C.W. Rock inhibition facilitates the generation of human-induced pluripotent stem cells in a defined, feeder-, and serum-free system. Cell Reprogram. 2010, 12, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Pan, W. Gsk3: A multifaceted kinase in Wnt signaling. Trends Biochem. Sci. 2010, 35, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Barrandon, O.; Nichols, J.; Kawaguchi, J.; Theunissen, T.W.; Smith, A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008, 6, e253. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhou, H.; Abujarour, R.; Zhu, S.; Young Joo, J.; Lin, T.; Hao, E.; Scholer, H.R.; Hayek, A.; Ding, S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells 2009, 27, 2992–3000. [Google Scholar] [PubMed]
- Lyssiotis, C.A.; Foreman, R.K.; Staerk, J.; Garcia, M.; Mathur, D.; Markoulaki, S.; Hanna, J.; Lairson, L.L.; Charette, B.D.; Bouchez, L.C.; et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc. Natl. Acad. Sci. USA 2009, 106, 8912–8917. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.D.; Klaus, A.; Garratt, A.N.; Birchmeier, W. Wnt signaling in stem and cancer stem cells. Curr. Opin. Cell Biol. 2013, 25, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Marson, A.; Foreman, R.; Chevalier, B.; Bilodeau, S.; Kahn, M.; Young, R.A.; Jaenisch, R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell 2008, 3, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Wilcoxen, K.M.; Nomoto, K.; Wu, S. Recent advances of mek inhibitors and their clinical progress. Curr. Top. Med. Chem. 2007, 7, 1364–1378. [Google Scholar] [PubMed]
- Telugu, B.P.; Ezashi, T.; Roberts, R.M. Porcine induced pluripotent stem cells analogous to naive and primed embryonic stem cells of the mouse. Int. J. Dev. Biol. 2010, 54, 1703–1711. [Google Scholar] [CrossRef] [PubMed]
- Varga, A.C.; Wrana, J.L. The disparate role of BMP in stem cell biology. Oncogene 2005, 24, 5713–5721. [Google Scholar] [CrossRef] [PubMed]
- Ichida, J.K.; Blanchard, J.; Lam, K.; Son, E.Y.; Chung, J.E.; Egli, D.; Loh, K.M.; Carter, A.C.; di Giorgio, F.P.; Koszka, K.; et al. A small-molecule inhibitor of TGF-β signaling replaces Sox2 in reprogramming by inducing nanog. Cell Stem Cell 2009, 5, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Maherali, N.; Hochedlinger, K. TGFβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009, 19, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.; Qian, C.; Tang, K.; Abd-Allah, S.M.; Jing, N. Inhibition of transforming growth factor β (TGF-β) signaling can substitute for Oct4 protein in reprogramming and maintain pluripotency. J. Biol. Chem. 2015, 290, 4500–4511. [Google Scholar] [CrossRef] [PubMed]
- Trokovic, R.; Weltner, J.; Manninen, T.; Mikkola, M.; Lundin, K.; Hamalainen, R.; Suomalainen, A.; Otonkoski, T. Small molecule inhibitors promote efficient generation of induced pluripotent stem cells from human skeletal myoblasts. Stem Cells Dev. 2013, 22, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, W.; Zhu, S.; Zhu, J.; Shi, Y.; Lin, T.; Hao, E.; Hayek, A.; Deng, H.; Ding, S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 2009, 4, 16–19. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, D.; Jiang, P. Chemically Induced Reprogramming of Somatic Cells to Pluripotent Stem Cells and Neural Cells. Int. J. Mol. Sci. 2016, 17, 226. https://doi.org/10.3390/ijms17020226
Biswas D, Jiang P. Chemically Induced Reprogramming of Somatic Cells to Pluripotent Stem Cells and Neural Cells. International Journal of Molecular Sciences. 2016; 17(2):226. https://doi.org/10.3390/ijms17020226
Chicago/Turabian StyleBiswas, Dhruba, and Peng Jiang. 2016. "Chemically Induced Reprogramming of Somatic Cells to Pluripotent Stem Cells and Neural Cells" International Journal of Molecular Sciences 17, no. 2: 226. https://doi.org/10.3390/ijms17020226