Role of Epigenetics in Stem Cell Proliferation and Differentiation: Implications for Treating Neurodegenerative Diseases
Abstract
:1. Introduction
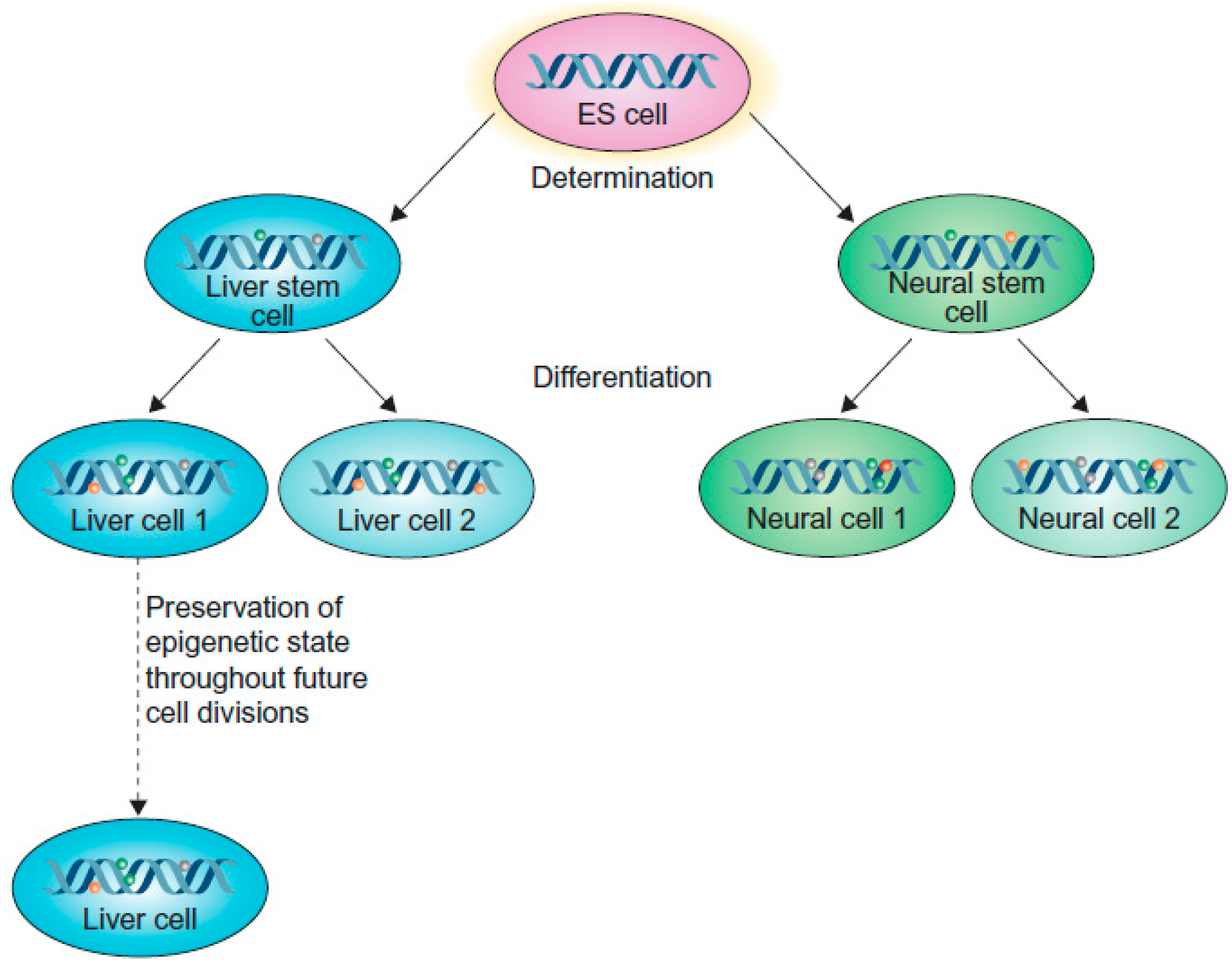
2. Epigenetics Regulate Cell Differentiation

| Molecules Regulate Gene Expression | Types of Modification | Examples | Outcomes |
|---|---|---|---|
| Histones | Methylation [30] |  | Histone methylation commonly silences the gene by repressing transcription, involving factors such as H3K27me3 [26]. |
| Acetylation [31,32] |  | Histone acetylation activates genes by enhancing the transcription, involving factors such as H3K4ac [31]. | |
| Phosphorylation [33,34] |  | Histone phosphorylation helps in chromatin remodeling and repairs the damaged DNA, such as H3T6 phosphorylation [35]. |
| Molecules that Regulate Gene Expression | Types of Modification | Examples | Outcome |
|---|---|---|---|
| Micro-RNA | Methylation | miR9 | Associated with cancer metastasis by gene repression [37]. |
| miR-34b/c | Affects the gene expression of miR-34: miR-34a, miR-34b and miR-34c; and it is associated with colorectal cancer [38]. | ||
| miR-124 | Associated with brain tumor by transcription repression [39]. |
| Molecules Regulate Gene Expression | Type of Modification | Examples | Outcomes |
|---|---|---|---|
| DNA-binding proteins | Methylation | Polycomb group proteins (PcG) | Play a role in cellular differentiation by repressing transcription [41]. |
| Heterochromatin protein (HP1) | Include many functions, like repressing genes by heterochromatin formation, regulates binding of complexes to centromere and maintains chromatin integrity [42]. | ||
| DNA binding zinc finger protein (ZnF) | Regulates transcription processes, such as C2H2 ZnFs [43]. |
3. Epigenetic Mechanisms in Mesenchymal Stem Cells and Huntington’s Disease
4. Epigenetic Mechanism in Adult Neural Stem Cells and Alzheimer’s Disease
4.1. DNA Methylation
4.2. Histone Modifications
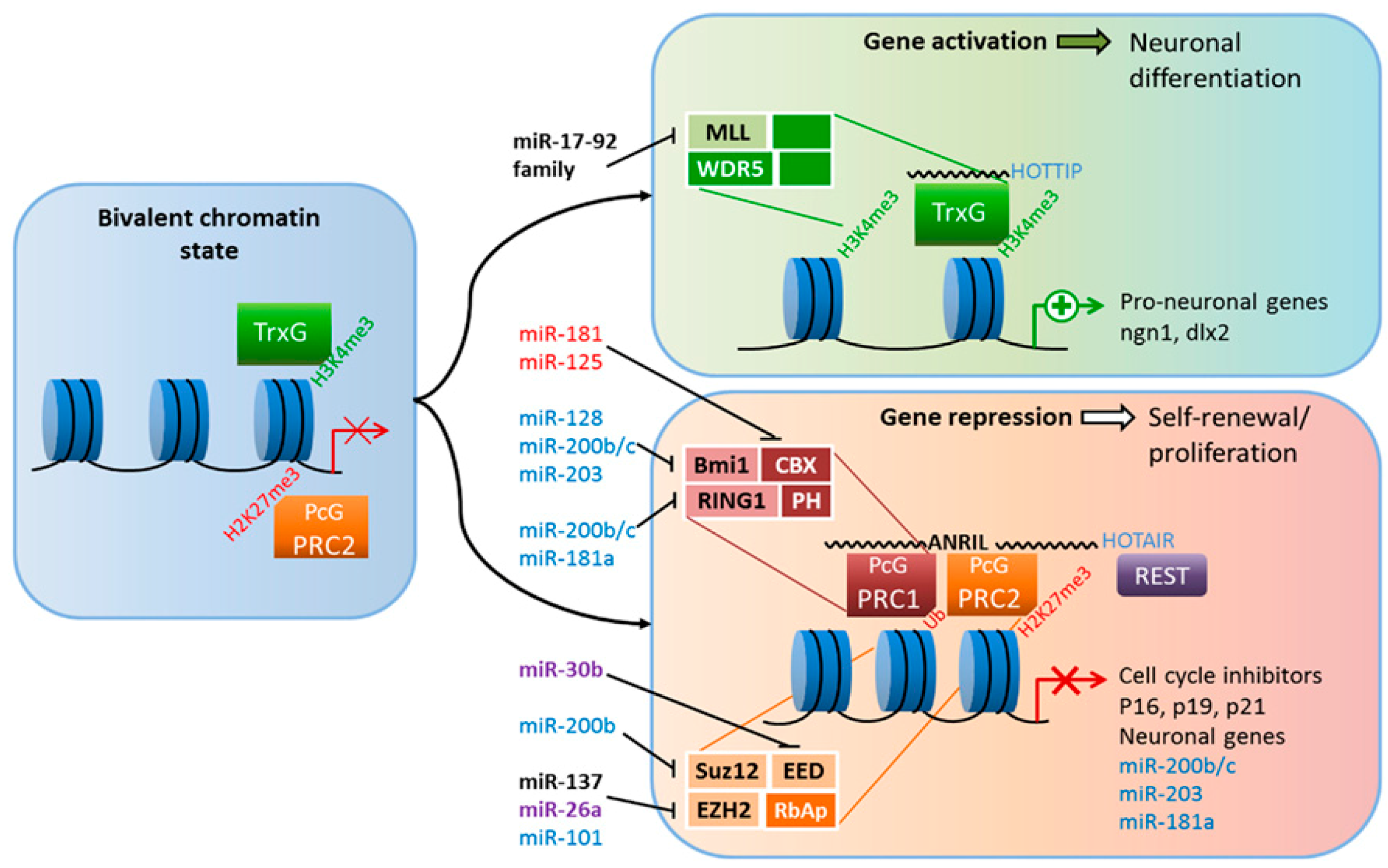
4.3. Micro-RNAs
| miRNAs | Gene Regulatory Mechanism(s) | Outcome(s) |
|---|---|---|
| miRNA-9 | Interacts with Tlx | Controls NSC neurogenesis |
| Let-b | Interacts with Tlx and cyclinD | Represses NSC proliferation and increases differentiation |
| miRNA-124 | Interacts with JAG1 | Induces neural progenitor renewal |
| Interacts with SOX9 | Controls glial cell renewal | |
| Interacts with DLX2 | Produces inter-neurons |
5. Epigenetic Mechanisms in Induced Pluripotent Stem Cells

| Genes of Induction | Outcome(s) in the Presence of the Factor | YamanakA Factors | Outcome(s) in the Presence of the Factor |
|---|---|---|---|
| Sox family (Sox1, Sox2, Sox3, and Sox15) | Mainly associated with maintaining the pluripotency of the cell. Functions of Sox 2 are dosage dependent. Associated with early embryonic development (tissues and organ formation) [82]. | Oct4 | Associated with pluripotency and silenced when cells undergo differentiation [87]. |
| Klf family (Klf1, Klf2, Klf4, and Klf5) | Associated with cell proliferation, differentiation and maintains tissue homeostasis and apoptosis [83]. | Sox2 | Associated with maintaining the embryonic stem cells in an undifferentiated state [82]. |
| Myc family (c-myc, L-myc, and N-myc) | Associated with tumor or cancer formation [84]. | Klf4 | They are required for reprogramming and self-renewal of embryonic stem cells [83]. |
| Nanog | Similar to Oct-3/4, they maintain pluripotency [85]. | c-Myc | Associated with early reprogramming and cell proliferation. They are also associated in the process of the transcriptional activity of some of the genes that undergo de-differentiation and proliferation [15]. |
| LIN28 | Associated with maintaining pluripotency by regulating miR let 7 [86]. |
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sell, S. Stem Cells Handbook, 2nd ed.; Humana Press: New York, NY, USA, 2013. [Google Scholar]
- Lakshmipathy, U.; Verfaillie, C. Stem cell plasticity. Blood Rev. 2005, 19, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zhang, F.; Xue, Q.; Ma, Z.; Lu, P.; Yu, B. Transplantation of bone marrow mesenchymal stem cells reduces lesion volume and induces axonal regrowth of injured spinal cord. Neuropathology 2010, 30, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Song, C.H.; Honmou, O.; Ohsawa, N.; Nakamura, K.; Hamada, H.; Furuoka, H.; Hasebe, R.; Horiuchi, M. Effect of Transplantation of Bone Marrow-Derived Mesenchymal Stem cells on Mice Infected with Prions. J. Virol. 2009, 83, 5918–5927. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J.; Gregory, C.A.; Spees, J.L. One Strategy for Cell and Gene Therapy: Harnessing the Power of Adult Stem Cells to Repair Tissues. Proc. Natl. Acad. Sci. USA 2003, 100, 11917–11923. [Google Scholar] [CrossRef] [PubMed]
- Blurton-Jones, M.; Spencer, B.; Michael, S.; Castello, N.A.; Agazaryan, A.A.; Davis, J.L.; Müller, F.J.; Loring, J.F.; Masliah, E.; LaFerla, F.M. Neural stem cells genetically-modified to express neprilysin reduce pathology in Alzheimer transgenic models. Stem Cell Res. Ther. 2014, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Crane, A.T.; Rossignol, J.; Dunbar, G.L. Use of Genetically Altered Stem Cells for the treatment of Huntington’s Disease. Brain Sci. 2014, 4, 202–219. [Google Scholar] [CrossRef] [PubMed]
- D’Anglemont de Tassigny, X.; Pascual, A.; López-Barneo, J. GDNF-based therapies, GDNF-producing interneurons, and trophic support of the dopaminergic nigrostriatal pathway. Implications for Parkinson’s disease. Front. Neuroanat. 2015, 9. [Google Scholar] [CrossRef]
- Wyse, R.D.; Dunbar, G.L.; Rossignol, J. Use of genetically modified mesenchymal stem cells to treat neurodegenerative diseases. Int. J. Mol. Sci. 2014, 15, 1719–1745. [Google Scholar] [CrossRef] [PubMed]
- Schu, S.; Nosov, M.; O’Flynn, L.; Shaw, G.; Treacy, O.; Barry, F.; Murphy, M.; O’Brien, T.; Ritter, T. Immunogenicity of Allogeneic Mesenchymal Stem Cells. J. Cell. Mol. Med. 2012, 16, 2094–2103. [Google Scholar] [CrossRef] [PubMed]
- Bifari, F.; Pacelli, L.; Krampera, M. Immunological properties of embryonic and adult stem cells. World J. Stem Cells. 2010, 2, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Boquest, A.C.; Noer, A.; Collas, P. Epigenetic programming of mesenchymal stem cells from human adipose tissue. Stem Cell. Rev. 2006, 2, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Ariff, I.M.; Mitra, A.; Basu, A. Epigenetic Regulation of Self-Renewal and Fate determination in Neural Stem Cells. J. Neurosci. Res. 2012, 90, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Lunyak, V.V.; Rosenfeld, M.G. Epigenetic regulation of stem cell fate. Hum. Mol. Genet. 2008, 17, R28–R36. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Plath, K. The roles of the reprogramming factors Oct4, Sox2 and Klf4 in resetting the somatic cell epigenome during induced pluripotent stem cell generation. Genome Biol. 2012, 13, 251. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010, 28, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Isles, A.R.; Wilkinson, L.S. Epigenetics: What is it and why is it important to mental disease. Br. Med. Bull. 2008, 85, 35–45. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, A.; Brickner, J.H. Mechanisms of epigenetic memory. Trends Genet. 2014, 30, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Sweatt, J.D.; Nestler, E.J.; Meaney, M.J.; Akbarian, S. An Overview of the Molecular Basis of Epigenetics. In Epigenetic Regulation in the Nervous System—Basic Mechanisms and Clinical Impact; Elsevier: San Diego, CA, USA, 2013; pp. 3–33. [Google Scholar]
- Bird, A.P. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. An Introduction to Modern Genetics. Proc. R. Entomol. Soc. Lond. Ser. A 1939, 14, 82. [Google Scholar]
- Stern, C.D.; Conrad, H. Waddington’s contributions to avian and mammalian development, 1930–1940. Int. J. Dev. Biol. 2000, 44, 15–22. [Google Scholar] [PubMed]
- Barth, T.K.; Imhof, A. Fast signals and slow marks: The dynamics of histone modifications. Trends Biochem. Sci. 2010, 35, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Zhang, Q.; Andersen, M.E. A deterministic map of Waddington’s epigenetic landscape for cell fate specification. BMC Syst. Biol. 2011, 5, 85. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Voigt, P.; Tee, W.W.; Reinberg, D. A double take on bivalent promoters. Genes Dev. 2013, 27, 1318–1338. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell. Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Delcuve, G.P.; Khan, D.H.; Davie, J.R. Roles of histone deacetylases in epigenetic regulation: Emerging paradigms from studies with inhibitors. Clin. Epigenet. 2012, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Hans, F.; Dimitrov, S. Histone H3 phosphorylation and cell division. Oncogene 2001, 20, 3021–3027. [Google Scholar] [CrossRef] [PubMed]
- Ghavifekr Fakhr, M.; Farshdousti Hagh, M.; Shanehbandi, D.; Baradaran, B. DNA methylation pattern as important epigenetic criterion in cancer. Genet. Res. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, B.; Drogaris, P.; Lin, H.H.; Armstrong, H.; Hiragami-Hamada, K.; Imhof, A.; Bonneil, E.; Thibault, P.; Verreault, A.; Festenstein, R.J. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011, 7, e1001354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UK Essays. Targeting The Prostate Cancer Epigenome Biology Essay. Available online: http://www.ukessays.com/essays/biology/targeting-the-prostate-cancer-epigenome-biology-essay.php?cref=1 (accessed on 19 December 2015).
- Kwon, S.J.; Park, J.H.; Park, E.J.; Lee, S.A.; Lee, H.S.; Kang, S.W.; Kwon, J. ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 2015, 34, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, T.; Chakravarti, D. A peek into the complex realm of histone phosphorylation. Mol. Cell. Biol. 2011, 31, 4858–4873. [Google Scholar] [CrossRef] [PubMed]
- Sawicka, A.; Seiser, C. Sensing core histone phosphorylation—A matter of perfect timing. Biochim. Biophys. Acta 2014, 1839, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Saetrom, P.; Snøve, O., Jr.; Rossi, J.J. Epigenetics and microRNAs. Pediatr. Res. 2007, 61, 17R–23R. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Zhu, Y.; Dai, Y.; Zeng, T.; Liu, L.; Li, J.; Wang, H.; Qin, Y.; Zeng, M.; et al. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget 2014, 5, 11669–11680. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zong, P.; Wang, W.; Liu, D.; Li, B.; Wang, Y.; Hu, J.; Ren, Y.; Qi, Y.; Cui, X.; et al. Hypermethylation of potential tumor suppressor miR-34b/c is correlated with late clinical stage in patients with soft tissue sarcomas. Exp. Mol. Pathol. 2015, 98, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Vijayakumarm, T.; Bakhshinyan, D.; Venugopal, C.; Singh, S.K. MicroRNA Regulation of Brain Tumour Initiating Cells in Central Nervous System Tumours. Stem Cells Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Bonifer, C.; Cockerill, P.N. Chromatin mechanisms regulating gene expression in health and disease. Adv. Exp. Med. Biol. 2011, 711, 12–25. [Google Scholar] [PubMed]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.T.; Bender, W.; Kingston, R.E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef]
- Norwood, L.E.; Grade, S.K.; Cryderman, D.E.; Hines, K.A.; Furiasse, N.; Toro, R.; Li, Y.; Dhasarathy, A.; Kladde, M.P.; Hendrix, M.J.; et al. Conserved properties of HP1(Hsalpha). Gene 2004, 336, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Najafabadi, H.S.; Mnaimneh, S.; Schmitges, F.W.; Garton, M.; Lam, K.N.; Yang, A.; Albu, M.; Weirauch, M.T.; Radovani, E.; Kim, P.M.; et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015, 33, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Archer, T. Epigenetic Changes Induced by Exercise. J. Reward Defic. Syndr. 2015, 1, 71–74. [Google Scholar]
- Zuccato, C.; Valenza, M.; Cattaneo, E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 2010, 90, 905–981. [Google Scholar] [CrossRef] [PubMed]
- HD Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar]
- Valtieri, M.; Sorrentino, A. The mesenchymal stromal cell contribution to homeostasis. J. Cell. Physiol. 2008, 217, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Wagner, W.; Feldmann, R.E., Jr.; Seckinger, A.; Maurer, M.H.; Wein, F.; Blake, J.; Krause, U.; Kalenka, A.; Bürgers, H.F.; Saffrich, R.; et al. The heterogeneity of human mesenchymal stem cell preparations--Evidence from simultaneous analysis of proteomes and transcriptomes. Exp. Hematol. 2006, 34, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Phinney, D.G. Functional heterogeneity of mesenchymal stem cells: Implications for cell therapy. J. Cell. Biochem. 2012, 113, 2806–2812. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, C.; Xie, Z.; Song, P.; Zhao, R.C.H.; Guo, L.; Liu, Z.; Wu, Y. Epigenetic Dysregulation in Mesenchymal Stem Cell Aging and Spontaneous Differentiation. PLoS ONE 2011, 6, e20526. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhou, Y.; Wang, S.; Wu, Y. Epigenetic changes of mesenchymal stem cells in three-dimensional (3D) spheroids. Cell. Mol. Med. 2014, 18, 2009–2019. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, J.; Fink, K.D.; Crane, A.T.; Davis, K.K.; Bombard, M.C.; Clerc, S.; Bavar, A.M.; Lowrance, S.A.; Song, C.; Witte, S.; et al. Reductions in behavioral deficits and neuropathology in the R6/2 mouse model of Huntington’s disease following transplantation of bone-marrow-derived mesenchymal stem cells is dependent on passage number. Stem Cell Res. Ther. 2015, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Teven, C.M.; Liu, X.; Hu, N.; Tang, N.; Kim, S.H.; Huang, E.; Yang, K.; Li, M.; Gao, J.L.; Liu, H.; et al. Epigenetic regulation of mesenchymal stem cells: A focus on osteogenic and adipogenic differentiation. Stem Cells Int. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Li, G.; Jiang, X.H. Fate determination in mesenchymal stem cells: A perspective from histone-modifying enzymes. Stem Cell. Res. Ther. 2015, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.M.; Selkoe, D.J. Oligomers on the brain: The emerging role of soluble protein aggregates in neurodegeneration. Protein Pept. Lett. 2004, 11, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. The origins of Alzheimer disease: A is for amyloid. JAMA 2000, 283, 1615–1617. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B. Loss-of-function presenilin mutations in Alzheimer disease. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007, 8, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Wagers, A.J. No place like home: Anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 2008, 9, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Lazarov, O.; Marr, R.A. Neurogenesis and Alzheimer’s disease: At the crossroads. Exp. Neurol. 2010, 223, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Fitzsimons, C.P.; van Bodegraven, E.; Schouten, M.; Lardenoije, R.; Kompotis, K.; Kenis, G.; van den Hurk, M.; Boks, M.P.; Biojone, C.; Joca, S.; et al. Epigenetic regulation of adult neural stem cells: Implications for Alzheimer’s disease. Mol. Neurodegener. 2014, 9, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldwell, M.A.; He, X.; Wilkie, N.; Pollack, S.; Marshall, G.; Wafford, K.A.; Svendsen, C.N. Growth factors regulate the survival and fate of cells derived from human neurospheres. Nat. Biotechnol. 2001, 19, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Shiue, K.; Schomberg, D.; Zhou, F. Cellular epigenetic modifications of neural stem cell differentiation. Cell Transplant. 2009, 18, 1197–1211. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.E.; Ebers, G.C.; Ramagopalan, S.V. Epigenetics: Molecular mechanisms and implications for disease. Trends Mol. Med. 2010, 16, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Jawerka, M.; Colak, D.; Dimou, L.; Spiller, C.; Lagger, S.; Montgomery, R.L.; Olson, E.N.; Wurst, W.; Göttlicher, M.; Götz, M. The specific role of histone deacetylase 2 in adult neurogenesis. Neuron Glia Biol. 2010, 6, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Dalgard, C.L.; Wynder, C.; Doughty, M.L. Histone deacetylase inhibitors SAHA and sodium butyrate block G1-to-S cell cycle progression in neurosphere formation by adult subventricular cells. BMC Neurosci. 2011, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Nakashima, K.; Kuwabara, T.; Mejia, E.; Gage, F.H. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 16659–16664. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Eisch, A.J. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: Unraveling the genome to understand the mind. Neurobiol. Dis. 2011, 39, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Jobe, E.M.; McQuate, A.L.; Zhao, X. Crosstalk among Epigenetic Pathways Regulates Neurogenesis. Front. Neurosci. 2012, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sun, G.; Li, S.; Lang, M.-F.; Yang, S.; Li, W.; Shi, Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 1876–1881. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Brohede, J.; Rinde, M.; Winblad, B.; Graff, C. A DNA methylation study of the amyloid precursor protein gene in several brain regions from patients with familial Alzheimer disease. J. Neurogenet. 2010, 24, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Barrachina, M.; Ferrer, I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J. Neuropathol. Exp. Neurol. 2009, 68, 880–891. [Google Scholar] [CrossRef] [PubMed]
- West, R.L.; Lee, J.M.; Maroun, L.E. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J. Mol. Neurosci. 1995, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Femminella, G.D.; Ferrara, N.; Rengo, G. The emerging role of microRNAs in Alzheimer’s disease. Front. Physiol. 2015, 6, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bicchi, I.; Morena, F.; Montesano, S.; Polidoro, M.; Martino, S. MicroRNAs and Molecular Mechanisms of Neurodegeneration. Genes 2013, 4, 244–263. [Google Scholar] [CrossRef] [PubMed]
- Maciotta, S.; Meregalli, M.; Torrente, Y. The involvement of microRNAs in neurodegenerative diseases. Front. Cell. Neurosci. 2013, 7, 265. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induced pluripotent stem cells in medicine and biology. Development 2013, 140, 2457–2461. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaskova, E.A.; Stekleneva, A.E.; Medvedev, S.P.; Zakian, S.M. “Epigenetic memory” phenomenon in induced pluripotent stem cells. Acta Nat. 2013, 5, 15–21. [Google Scholar]
- Stadtfeld, M.; Hochedlinger, K. Induced pluripotency: History, mechanisms, and applications. Genes Dev. 2010, 24, 2239–2263. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Hirano, K.; Nagata, S.; Tada, T. Sox2 expression effects on direct reprogramming efficiency as determined by alternative somatic cell fate. Stem Cells Res. 2011, 6, 177–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Ai, W. Function of KLF4 in Stem Cell Biology. In Pluripotent Stem Cells; Bhartiya, D., Ed.; Intech: Rijeka, Croatia, 2013. [Google Scholar]
- Tansey, W.P. Mammalian MYC Proteins and Cancer. N. J. Sci. 2014, 2014. [Google Scholar] [CrossRef]
- Baker, M. What does Nanog do? Nat. Rep. Stem Cells 2009. [Google Scholar] [CrossRef]
- Tsialikas, J.; Romer-Seibert, J. LIN28: Roles and regulation in development and beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Jin, Y. Role of Oct4 in maintaining and regaining stem cell pluripotency. Stem Cell Res. Ther. 2010, 1, 39. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Toyoda, M.; Yamazaki-Inoue, M.; Fukawatase, Y.; Chikazawa, E.; Sakaguchi, H.; Akutsu, H.; Umezawa, A. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011, 9, e153–e160. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.; Koch, C.; Gupta, M.K.; Lin, Q.; Lenz, M.; Laufs, S.; Denecke, B.; Schmidt, M.; Linke, M.; Hennies, H.C.; et al. Induced pluripotent mesenchymal stromal cell clones retain donor-derived differences in DNA methylation profiles. Mol. Ther. 2013, 21, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Papp, B.; Plath, K. Epigenetics of reprogramming to induced pluripotency. Cell 2013, 152, 1324–1343. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srinageshwar, B.; Maiti, P.; Dunbar, G.L.; Rossignol, J. Role of Epigenetics in Stem Cell Proliferation and Differentiation: Implications for Treating Neurodegenerative Diseases. Int. J. Mol. Sci. 2016, 17, 199. https://doi.org/10.3390/ijms17020199
Srinageshwar B, Maiti P, Dunbar GL, Rossignol J. Role of Epigenetics in Stem Cell Proliferation and Differentiation: Implications for Treating Neurodegenerative Diseases. International Journal of Molecular Sciences. 2016; 17(2):199. https://doi.org/10.3390/ijms17020199
Chicago/Turabian StyleSrinageshwar, Bhairavi, Panchanan Maiti, Gary L. Dunbar, and Julien Rossignol. 2016. "Role of Epigenetics in Stem Cell Proliferation and Differentiation: Implications for Treating Neurodegenerative Diseases" International Journal of Molecular Sciences 17, no. 2: 199. https://doi.org/10.3390/ijms17020199








