Transcriptome-Wide Identification and Expression Profiling Analysis of Chrysanthemum Trihelix Transcription Factors
Abstract
:1. Introduction
2. Results
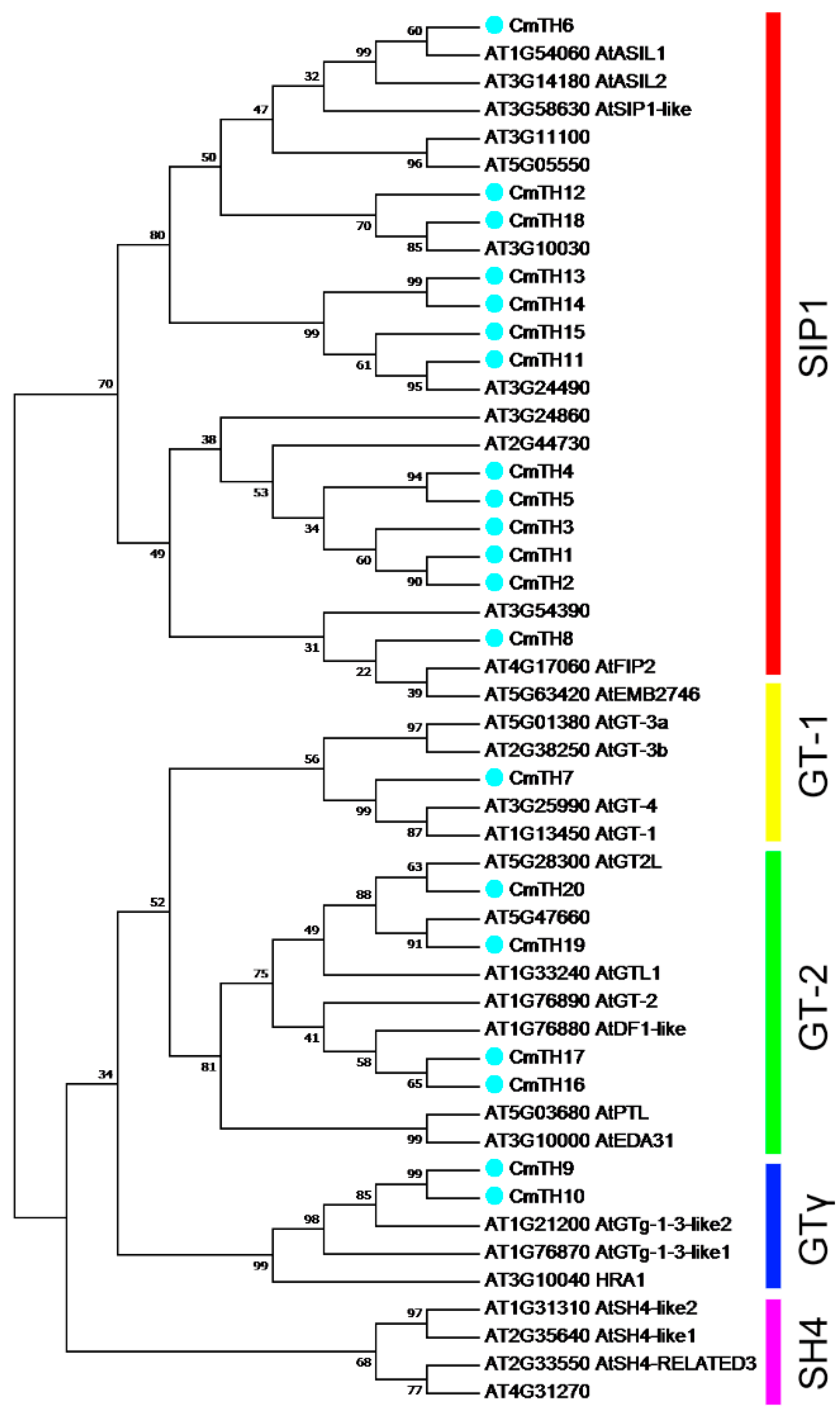
2.1. Identification and Phylogenetic Analysis of Putative Trihelix Factors in Chrysanthemum
| Gene | GenBank Accession No. | Amino Acids Length (aa) | AtTH Orthologs | Locus Name | pI | MW | Subcellular Loclization |
|---|---|---|---|---|---|---|---|
| CmTH1 | KT253111 | 340 | AdMyb/SANT | AT2G44730.1 | 9.51 | 38,618.98 | nucleus |
| CmTH2 | KT253112 | 314 | AdMyb/SANT | AT2G44730.1 | 9.09 | 35,364.01 | nucleus |
| CmTH3 | KT253113 | 338 | AdMyb/SANT | AT2G44730.1 | 6.13 | 38,292.92 | nucleus |
| CmTH4 | KT253114 | 224 | AdMyb/SANT | AT2G44730.1 | 9.56 | 24,572.87 | chloroplast stroma |
| CmTH5 | KT253115 | 295 | AdMyb/SANT | AT2G44730.1 | 8.52 | 33,510.27 | nucleus |
| CmTH6 | KT253116 | 422 | ASIL2 | AT3G14180.1 | 9.27 | 47,237.56 | chloroplast thylakoid space |
| CmTH7 | KT253117 | 399 | HLP | AT3G25990.1 | 6.85 | 45,568.92 | microbody |
| CmTH8 | KT253118 | 249 | SSDB-TF | AT3G54390.1 | 9.30 | 27,119.85 | nucleus |
| CmTH9 | KT253119 | 446 | SSDB-TF | AT1G21200.1 | 6.35 | 51,562.20 | cytoplasm |
| CmTH10 | KT253120 | 384 | SSDB-TF | AT1G21200.1 | 6.26 | 43,610.86 | cytoplasm |
| CmTH11 | KT253121 | 357 | AdMyb/SANT | AT3G24490.1 | 4.53 | 41,753.36 | mitochondrial matrix space |
| CmTH12 | KT253122 | 216 | SSDB-TF | AT5G05550.2 | 9.18 | 24,860.42 | cytoplasm |
| CmTH13 | KT253123 | 373 | AdMyb/SANT | AT3G24490.1 | 5.04 | 42,980.38 | nucleus |
| CmTH14 | KT253124 | 372 | AdMyb/SANT | AT3G24490.1 | 5.00 | 42,911.32 | nucleus |
| CmTH15 | KT253125 | 382 | AdMyb/SANT | AT3G24490.1 | 4.89 | 43,488.77 | nucleus |
| CmTH16 | KT253126 | 684 | DHLP | AT1G76880.1 | 6.68 | 76,768.06 | nucleus |
| CmTH17 | KT253127 | 597 | DHLP | AT1G76880.1 | 5.31 | 66,479.62 | nucleus |
| CmTH18 | KT253128 | 211 | SSDB-TF | AT5G05550.2 | 5.1 | 23,338.93 | cytoplasm |
| CmTH19 | KT253129 | 341 | HLP | AT5G47660.1 | 9.62 | 39,351.70 | nucleus |
| CmTH20 | KT253130 | 526 | DHLP | AT5G28300.1 | 5.75 | 61,139.47 | nucleus |
2.2. Conserved Sequences in TH Proteins
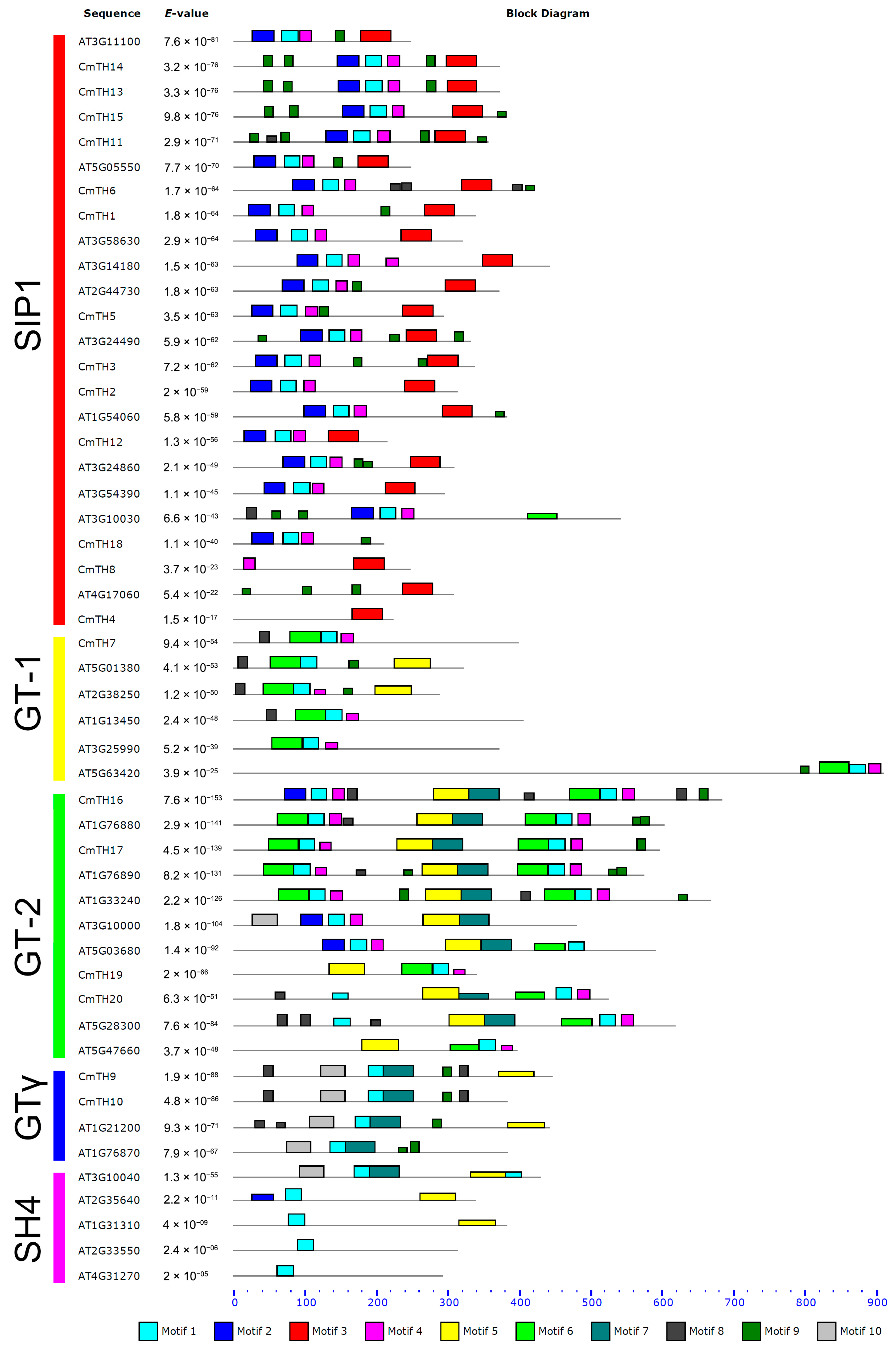
2.3. miRNA Target Site Prediction
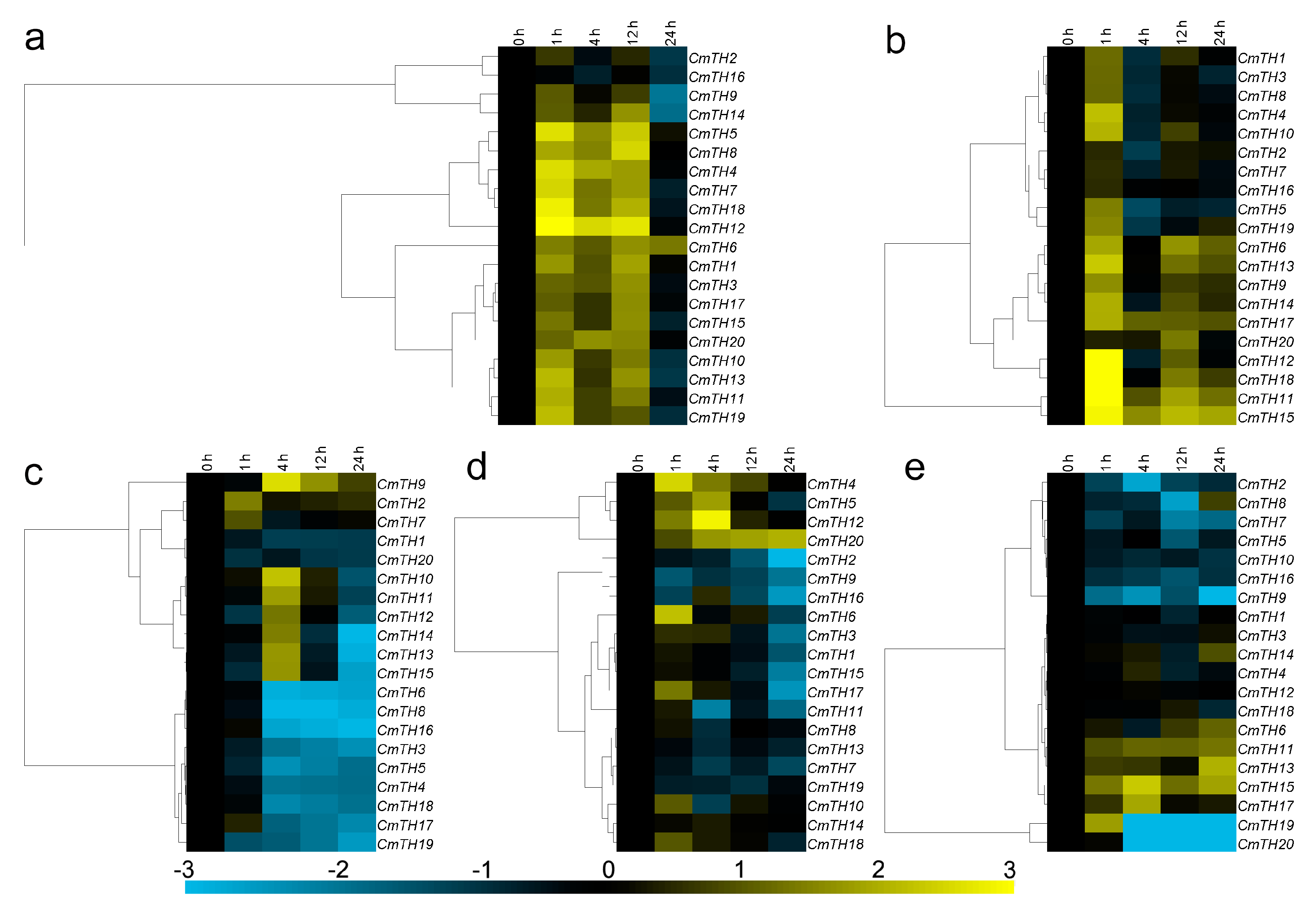
2.4. Transcription Profiling of CmTH Genes

2.5. Expression of CmTH Genes after Treatment with Phytohormones

2.6. Expression Profiling of CmTH Genes under Abiotic Stress
3. Discussion
3.1. The Trihelix Family in Chrysanthemum
3.2. Diverse Motifs Predicted in TH Factors
3.3. miRNA Target Site Prediction
3.4. Organ-Preferential Expression of CmTH Genes
3.5. Transcriptional Responses of Chrysanthemum Trihelix Genes after Phytohormone or against Abiotic Stress Treatment
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Database Searches and Sequencing of Full-Length CmTH cDNAs
4.3. Phylogenetic Tree Construction and Sequence Analysis
4.4. Plant Treatments
4.5. Real-Time Quantitative PCR (qPCR)
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Riechmann, J.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.; Samaha, R. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors—Light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Shan, H.; Chen, S.; Jiang, J.; Gu, C.; Zhou, G.; Chen, Y.; Song, A.; Chen, F. The heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB1 alters lignin composition and represses flavonoid synthesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e65680. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hong, G.; Han, B. Transcript abundance of rml1, encoding a putative GT1-like factor in rice, is up-regulated by Magnaporthe grisea and down-regulated by light. Gene 2004, 324, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Li, Q.T.; Chen, H.W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Trihelix transcription factor GT-4 mediates salt tolerance via interaction with TEM2 in Arabidopsis. BMC Plant Biol. 2014, 14, 339. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y. Several features of the GT-factor trihelix domain resemble those of the Myb DNA-binding domain. Plant Physiol. 2000, 124, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R.M.; Caspar, T.; Dehesh, K.; Quail, P.H. DNA binding factor GT-2 from Arabidopsis. Plant Mol. Biol. 1993, 23, 337–348. [Google Scholar] [CrossRef]
- Zhou, D.-X. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999, 4, 210–214. [Google Scholar] [CrossRef]
- Xie, Z.-M.; Zou, H.-F.; Lei, G.; Wei, W.; Zhou, Q.-Y.; Niu, C.-F.; Liao, Y.; Tian, A.-G.; Ma, B.; Zhang, W.-K. Soybean trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS ONE 2009, 4, e6898. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qin, G.; Chen, Z.; Gu, H.; Qu, L.-J. A gain-of-function mutation of transcriptional factor PTL results in curly leaves, dwarfism and male sterility by affecting auxin homeostasis. Plant Mol. Biol. 2008, 66, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Suprunova, T.; Krugman, T.; Distelfeld, A.; Fahima, T.; Nevo, E.; Korol, A. Identification of a novel gene (Hsdr4) involved in water-stress tolerance in wild barley. Plant Mol. Biol. 2007, 64, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Perisic, O.; Lam, E. A tobacco DNA binding protein that interacts with a light-responsive box II element. Plant Cell 1992, 4, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Yoo, C.Y.; Gosney, M.J.; Hasegawa, P.M.; Mickelbart, M.V. Poplar GTL1 is a Ca2+/calmodulin-binding transcription factor that functions in plant water use efficiency and drought tolerance. PLoS ONE 2012, 7, e32925. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lu, D.; Li, C.; Luo, J.; Zhu, B.-F.; Zhu, J.; Shangguan, Y.; Wang, Z.; Sang, T.; Zhou, B. Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 2012, 24, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Kuromori, T.; Wada, T.; Kamiya, A.; Yuguchi, M.; Yokouchi, T.; Imura, Y.; Takabe, H.; Sakurai, T.; Akiyama, K.; Hirayama, T. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006, 47, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; You, Y.; Chen, F.; Li, P.; Jiang, J.; Chen, S. A multiplex RT-PCR for rapid and simultaneous detection of viruses and viroids in chrysanthemum. Lett. Appl. Microbiol. 2013, 56, 8–13. [Google Scholar] [CrossRef]
- Vos, I.A.; Moritz, L.; Pieterse, C.M.; van Wees, S.C. Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 2015, 6, 639. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.-H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y.; Inaba, T.; Furuhashi, H.; Sasaki, Y. Trihelix DNA-binding protein with specificities for two distinctcis-elements both important for light down-regulated and dark-inducible gene expression in higher plants. J. Biol. Chem. 2001, 276, 22238–22243. [Google Scholar] [CrossRef] [PubMed]
- Barr, M.S.; Willmann, M.R.; Jenik, P.D. Is there a role for trihelix transcription factors in embryo maturation? Plant Signal. Behav. 2012, 7, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Niyada, E.; Fujimoto, N.; Nagasaki, Y.; Noto, K.; Miyanoiri, Y.; Murata, J.; Hiratsuka, K.; Katahira, M. Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: Mechanism for an increase in DNA-binding affinity through phosphorylation. Proteins 2010, 78, 3033–3047. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Cai, X.; Ye, Z.; Li, H. Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem. Biophys. Res. Commun. 2015, 468, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Murata, J.; Takase, H.; Hiratsuka, K. Characterization of a novel GT-box binding protein from Arabidopsis. Plant Biotechnol. 2002, 19, 103–112. [Google Scholar] [CrossRef]
- Breuer, C.; Kawamura, A.; Ichikawa, T.; Tominaga-Wada, R.; Wada, T.; Kondou, Y.; Muto, S.; Matsui, M.; Sugimoto, K. The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 2009, 21, 2307–2322. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.Y.; Pence, H.E.; Jin, J.B.; Miura, K.; Gosney, M.J.; Hasegawa, P.M.; Mickelbart, M.V. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 2010, 22, 4128–4141. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xie, K.; Hou, X.; Hu, H.; Xiong, L. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol. Genet. Genom. 2010, 283, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Schölkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Kitakura, S.; Fujita, T.; Ueno, Y.; Terakura, S.; Wabiko, H.; Machida, Y. The protein encoded by oncogene 6b from Agrobacterium tumefaciens interacts with a nuclear protein of tobacco. Plant Cell 2002, 14, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.-J.; Lydiate, D.J.; Li, X.; Lui, H.; Gjetvaj, B.; Hegedus, D.D.; Rozwadowski, K. Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 2009, 21, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, Z.; Dong, W.; Sun, C.; Wang, H.; Song, A.; He, L.; Fang, W.; Chen, F.; Teng, N. Transcriptomic and proteomic analysis reveals mechanisms of embryo abortion during chrysanthemum cross breeding. Sci. Rep. 2014, 4, 6536. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York City, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.; Chang, H.-Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Lu, J.; Jiang, J.; Chen, S.; Guan, Z.; Fang, W.; Chen, F. Isolation and characterisation of Chrysanthemum crassum SOS1, encoding a putative plasma membrane Na+/H+ antiporter. Plant Biol. 2012, 14, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; An, J.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 2014, 80, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Zhu, X.; Chen, F.; Gao, H.; Jiang, J.; Chen, S. A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int. J. Mol. Sci. 2014, 15, 5063–5078. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Li, P.; Jiang, J.; Chen, S.; Li, H.; Zeng, J.; Shao, Y.; Zhu, L.; Zhang, Z.; Chen, F. Phylogenetic and transcription analysis of chrysanthemum WRKY transcription factors. Int. J. Mol. Sci. 2014, 15, 14442–14455. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- De Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, A.; Wu, D.; Fan, Q.; Tian, C.; Chen, S.; Guan, Z.; Xin, J.; Zhao, K.; Chen, F. Transcriptome-Wide Identification and Expression Profiling Analysis of Chrysanthemum Trihelix Transcription Factors. Int. J. Mol. Sci. 2016, 17, 198. https://doi.org/10.3390/ijms17020198
Song A, Wu D, Fan Q, Tian C, Chen S, Guan Z, Xin J, Zhao K, Chen F. Transcriptome-Wide Identification and Expression Profiling Analysis of Chrysanthemum Trihelix Transcription Factors. International Journal of Molecular Sciences. 2016; 17(2):198. https://doi.org/10.3390/ijms17020198
Chicago/Turabian StyleSong, Aiping, Dan Wu, Qingqing Fan, Chang Tian, Sumei Chen, Zhiyong Guan, Jingjing Xin, Kunkun Zhao, and Fadi Chen. 2016. "Transcriptome-Wide Identification and Expression Profiling Analysis of Chrysanthemum Trihelix Transcription Factors" International Journal of Molecular Sciences 17, no. 2: 198. https://doi.org/10.3390/ijms17020198





