Identification and Expression Analysis of Polygalacturonase Family Members during Peach Fruit Softening
Abstract
:1. Introduction
2. Results
2.1. Identification of Polygalacturonase (PG) Family Members in Peach
2.2. Phylogenetic Analysis of PG Family Members in Peach
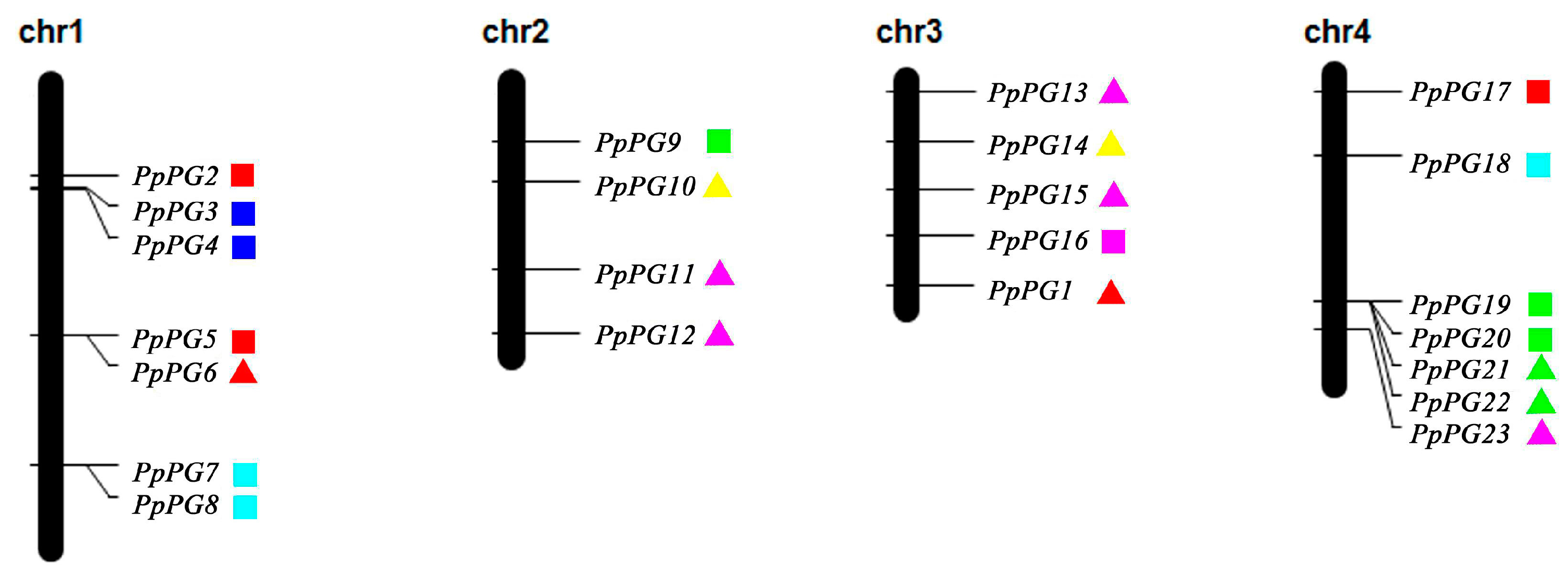
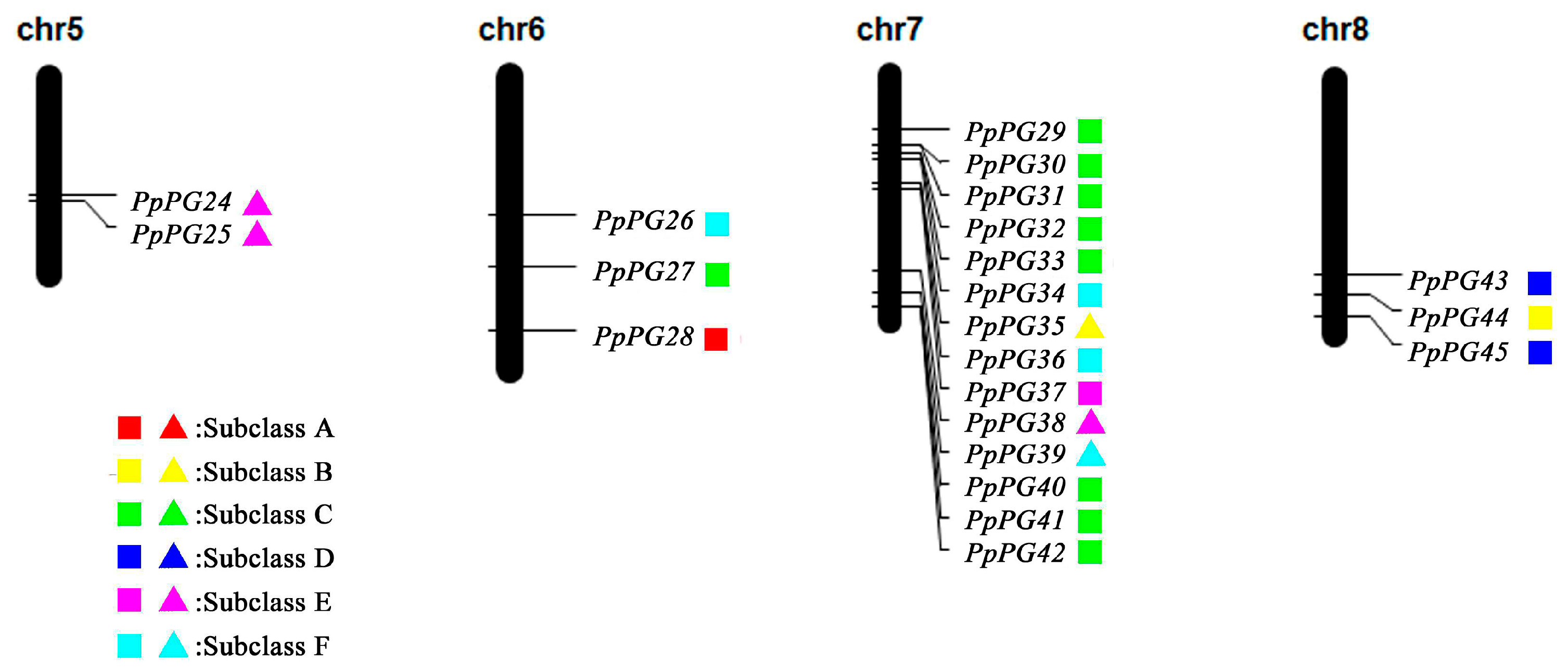
2.3. Genome Distribution and Gene Structures of PG Family Members in Peach
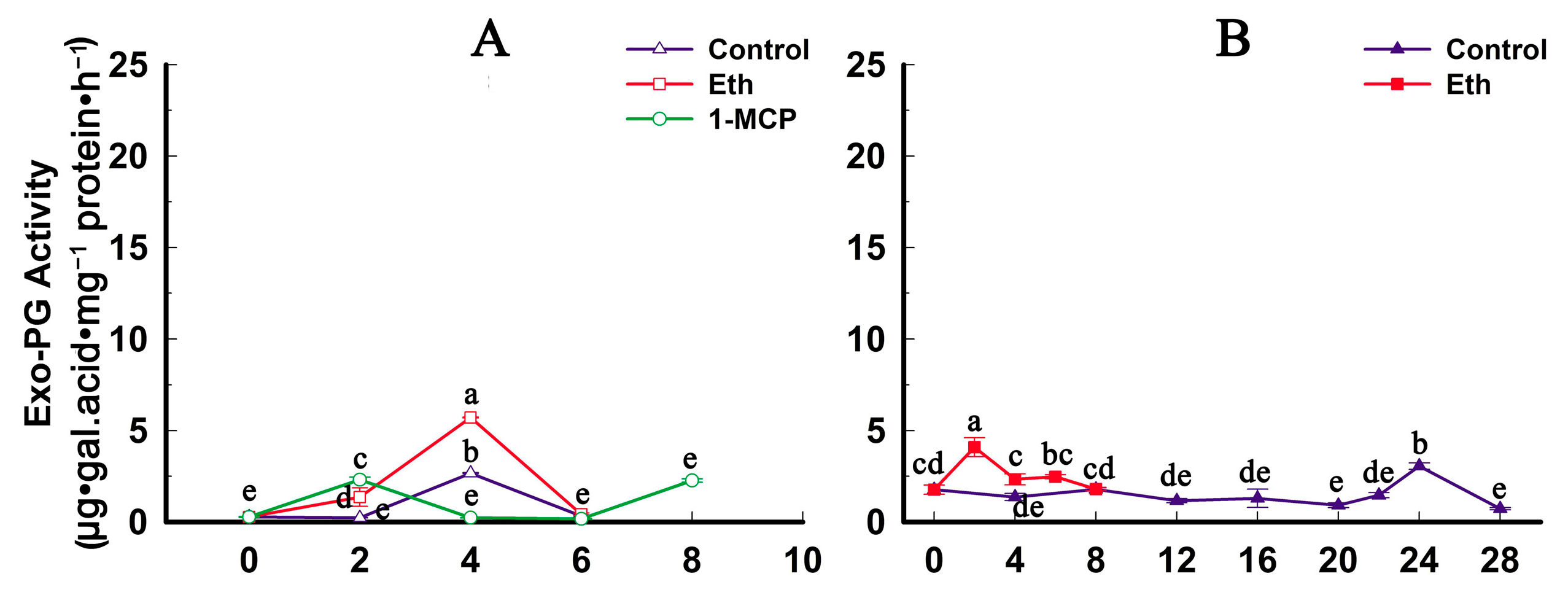
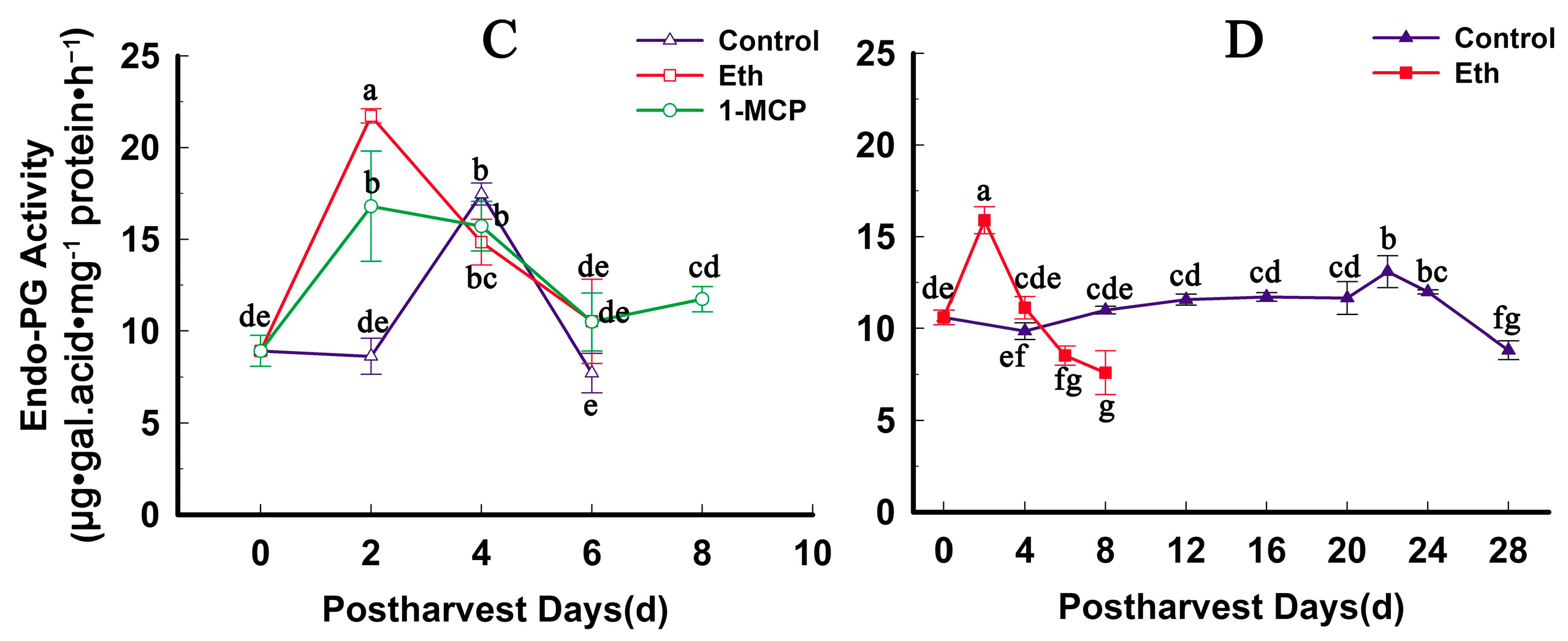
2.4. Fruit Firmness, Ethylene Production and PG Activity Change during Softening
2.5. Identification of PG Genes Exhibiting Ripening-Associated Patterns of Expression
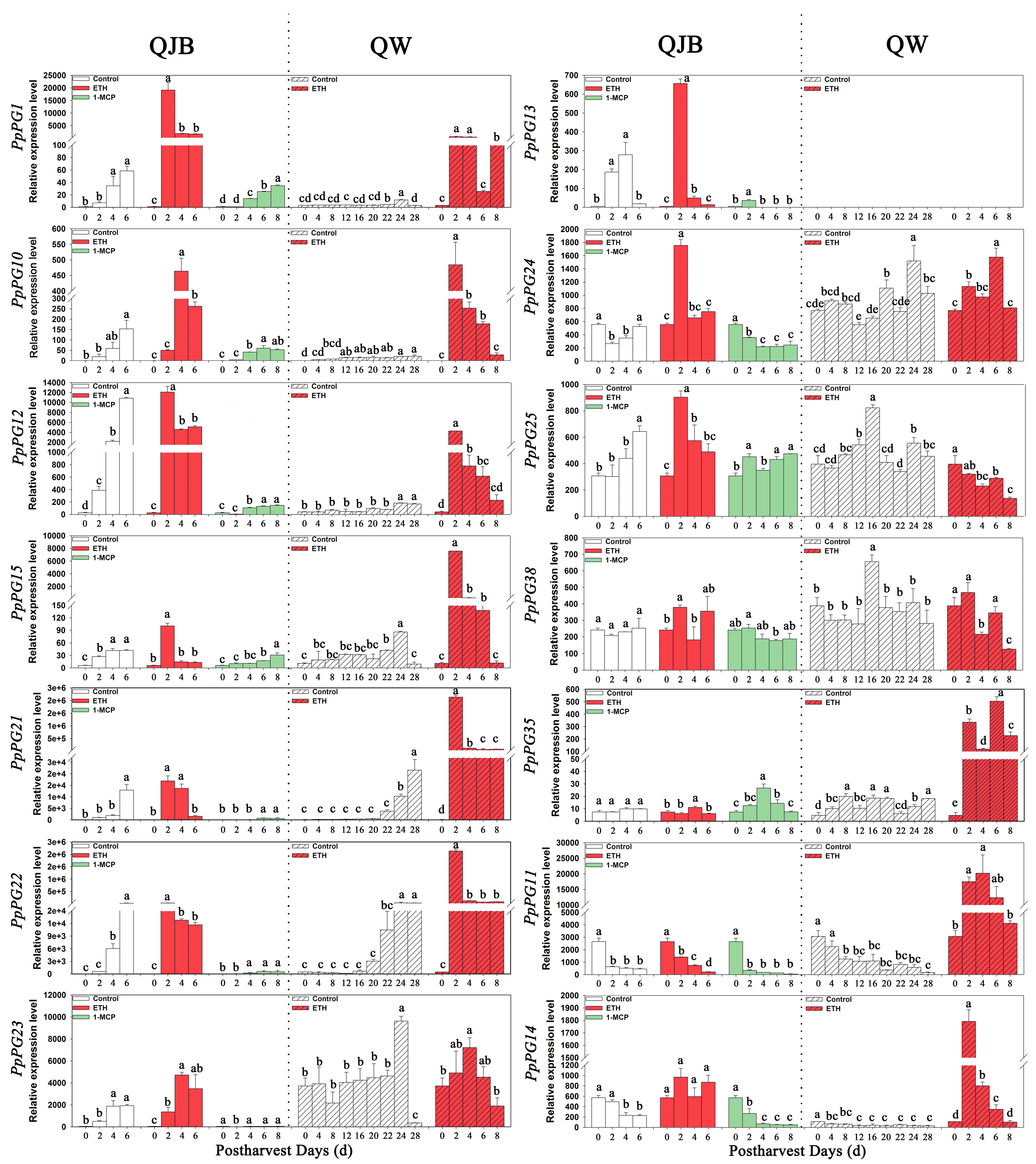
2.6. Ethylene and 1-MCP Alter PG Gene Expression
3. Discussion
3.1. PG Family Member Identification and Sequence Analysis
3.2. Possible Roles of PG Family Members in Fruit Softening during Ripening
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. RNA Extraction and Reverse Transcription
4.3. Identification of Peach PG Family Members
4.4. Multiple Sequence Alignment, Phylogenetic Analysis, Exon/Intron Structure Determination and Chromosome Locations
4.5. RNA Deep Sequencing and Library Construction
4.6. Real-Time Quantitative PCR (qPCR) Assays
4.7. Flesh Firmness, Ethylene Production and Determination of Enzyme Activity
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barry, C.S.; Giovannoni, J.J. Ethylene and fruit ripening. J. Plant Growth Regul. 2007, 26, 143–159. [Google Scholar] [CrossRef]
- Osorio, S.; Scossa, F.; Fernie, A.R. Molecular regulation of fruit ripening. Front. Plant Sci. 2013, 4, 198. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, H.; Hayama, H.; Tatsuki, M.; Nakamura, Y. Cell wall modification during development of mealy texture in the stony-hard peach “odoroki” treated with propylene. Postharvest Biol. Technol. 2010, 55, 1–7. [Google Scholar] [CrossRef]
- Brummell, D.A.; Dal Cin, V.; Crisosto, C.H.; Labavitch, J.M. Cell wall metabolism during maturation, ripening and senescence of peach fruit. J. Exp. Bot. 2004, 55, 2029–2039. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Chen, H.; Hahn, M.G.; Mohnen, D.; Xu, Y. Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiol. 2010, 153, 1729–1746. [Google Scholar] [CrossRef] [PubMed]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, M.C.; Briggs, S.P.H.; Knox, J.P. Intercellular adhesion and cell separation in plants. Plant Cell Environ. 2003, 26, 977–989. [Google Scholar] [CrossRef]
- Fry, S.C. The Growing Plant Cell Wall: Chemical and Metabolic Analysis; John Wiley & Sons: New York, NY, USA, 1988; pp. 332–365. [Google Scholar]
- Habibi, Y.; Heyraud, A.; Mahrouz, M.; Vignon, M.R. Structural features of pectic polysaccharides from the skin of opuntia ficus-indica prickly pear fruits. Carbohydr. Res. 2004, 339, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Carranza, Z.H.; Elliott, K.A.; Roberts, J.A. Expression of polygalacturonases and evidence to support their role during cell separation processes in arabidopsis thaliana. J. Exp. Bot. 2007, 58, 3719–3730. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.G.; Schröder, R.; Hallett, I.C.; Cohen, D.; MacRae, E.A. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002, 129, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Pose, S.; Paniagua, C.; Cifuentes, M.; Blanco-Portales, R.; Quesada, M.A.; Mercado, J.A. Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J. Exp. Bot. 2013, 64, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Hiwasa, K.; Kinugasa, Y.; Amano, S.; Hashimoto, A.; Nakano, R.; Inaba, A.; Kubo, Y. Ethylene is required for both the initiation and progression of softening in pear. J. Exp. Bot. 2003, 54, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.H.; Nath, P. Expression of multiple forms of polygalacturonase gene during ripening in banana fruit. Plant Physiol. Biochem. 2005, 43, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Fabi, J.P.; Cordenunsi, B.R.; Seymour, G.B.; Lajolo, F.M.; do Nascimento, J.R. Molecular cloning and characterization of a ripening-induced polygalacturonase related to papaya fruit softening. Plant Physiol. Biochem. 2009, 47, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Abu-Goukh, A.-B.A.; Bashir, H.A. Changes in pectic enzymes and cellulase activity during guava fruit ripening. Food Chem. 2003, 83, 213–218. [Google Scholar] [CrossRef]
- Rao, G.U.; Paran, I. Polygalacturonase: A candidate gene for the soft flesh and deciduous fruit mutation in capsicum. Plant Mol. Biol. 2003, 51, 135–141. [Google Scholar] [PubMed]
- Costa, F.; Peace, C.P.; Stella, S.; Serra, S.; Musacchi, S.; Bazzani, M.; Sansavini, S.; Van de Weg, W.E. QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in apple (malus x domestica borkh.). J. Exp. Bot. 2010, 61, 3029–3039. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.J.S.; Watson, C.F.; Ray, J.; Bird, C.R.; Morris, P.C.; Schuch, W.; Grierson, D. Antisense rna inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature 1988, 334, 724–726. [Google Scholar] [CrossRef]
- Callahan, A.M.; Scorza, R.; Bassett, C.; Nickerson, M.; Abeles, F.B. Deletions in an endopolygalacturonase gene cluster correlate with non-melting flesh texture in peach. Funct. Plant Biol. 2004, 31, 159–168. [Google Scholar] [CrossRef]
- Ghiani, A.; Onelli, E.; Aina, R.; Cocucci, M.; Citterio, S. A comparative study of melting and non-melting flesh peach cultivars reveals that during fruit ripening endo-polygalacturonase (endo-PG) is mainly involved in pericarp textural changes, not in firmness reduction. J. Exp. Bot. 2011, 62, 4043–4054. [Google Scholar] [CrossRef] [PubMed]
- Peace, C.P.; Crisosto, C.H.; Gradziel, T.M. Endopolygalacturonase: A candidate gene for freestone and melting fleshin peach. Mol. Breed. 2005, 16, 21–31. [Google Scholar] [CrossRef]
- Gu, C.; Wang, L.; Wang, W.; Zhou, H.; Ma, B.; Zheng, H.; Fang, T.; Ogutu, C.; Vimolmangkang, S.; Han, Y. Copy number variation of a gene cluster encoding endopolygalacturonase mediates flesh texture and stone adhesion in peach. J. Exp. Bot. 2016, 67, 1993–2005. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Wu, J.; Lu, W.; Li, J. A polygalacturonase gene clustered into clade e involved in lychee fruitlet abscission. Sci. Hortic. 2013, 150, 244–250. [Google Scholar] [CrossRef]
- González-Carranza, Z.H.; Whitelaw, C.A.; Swarup, R.; Roberts, J.A. Temporal and spatial expression of a polygalacturonase during leaf and flower abscission in oilseed rape and arabidopsis. Plant Physiol. 2002, 128, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-Z.; Lu, F.; Imsabai, W.; Meir, S.; Reid, M.S. Silencing polygalacturonase expression inhibits tomato petiole abscission. J. Exp. Bot. 2008, 59, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Somerville, C.R. Tetrad pollen formation in quartet mutants of arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J. 1998, 15, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. Arabidopsis Dehiscence zone Polygalacturonase1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Shiu, S.-H.; Thoma, S.; Li, W.-H.; Patterson, S.E. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006, 7, R87. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.L.; Liu, H.J.; Wang, X.R.; Zeng, Q.Y. Molecular evolution and expression divergence of the populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol. 2013, 197, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liang, Y.; Lv, M.; Wu, J.; Lu, G.; Cao, J. Genome-wide identification and characterization of polygalacturonase genes in cucumis sativus and citrullus lanatus. Plant Physiol. Biochem. 2014, 74, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Morgutti, S.; Negrini, N.; Nocito, F.F.; Ghiani, A.; Bassi, D.; Cocucci, M. Changes in endopolygalacturonase levels and characterization of a putative endo-PG gene during fruit softening in peach genotypes with nonmelting and melting flesh fruit phenotypes. New Phytol. 2006, 171, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Lester, D.R.; Speirs, J.; Orr, G.; Brady, C.J. Endopolygalacturonase cdna isolation and its expression analysis in melting and nonmelting peach cultivars. Plant Physiol. 1994, 105, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Murayama, H.; Arikawa, M.; Sasaki, Y.; dal Cin, V.; Mitsuhashi, W.; Toyomasu, T. Effect of ethylene treatment on expression of polyuronide-modifying genes and solubilization of polyuronides during ripening in two peach cultivars having different softening characteristics. Postharvest Biol. Technol. 2009, 52, 196–201. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Osborne, E.; Poindexter, P.D.; Somerville, C.R. Microspore separation in the QUARTET3 mutants of arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 2003, 133, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Sitrit, Y.; Hadfield, K.A.; Bennett, A.B.; Bradford, K.J.; Downie, A.B. Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol. 1999, 121, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Initiative, T.I.P.G.; Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar]
- Park, K.C.; Kwon, S.J.; Kim, P.H.; Bureau, T.; Kim, N.S. Gene structure dynamics and divergence of the polygalacturonase gene family of plants and fungus. Genome 2008, 51, 30–40. [Google Scholar] [PubMed]
- Stratilová, E.; Mislovičová, D.; Kačuráková, M.; Machová, E.; Kolarová, N.; Markovič, O.; Jörnvall, H. The glycoprotein character of multiple forms of aspergillus polygalacturonase. J. Protein Chem. 1998, 17, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Roongsattham, P.; Morcillo, F.; Jantasuriyarat, C.; Pizot, M.; Moussu, S.; Jayaweera, D.; Collin, M.; Gonzalez-Carranza, Z.H.; Amblard, P.; Tregear, J.W.; et al. Temporal and spatial expression of polygalacturonase gene family members reveals divergent regulation during fleshy fruit ripening and abscission in the monocot species oil palm. BMC Plant Biol. 2012, 12, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iglesias-Fernándeza, R.; Matilla, A.J.; Rodríguez-Gacio, M.C.; Fernández-Otero, C.; Torre, F.D.L. The polygalacturonase gene PdPG1 is developmentally regulated in reproductive organs of Prunus domestica L. Subsp. insititia. Plant Sci. 2007, 172, 763–772. [Google Scholar] [CrossRef]
- Kalaitzis, P.; Koehler, S.M.; Tucker, M.L. Cloning of a tomato polygalacturonase expressed in abscission. Plant Mol. Biol. 1995, 28, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Kalaitzis, P.; Solomos, T.; Tucker, M.L. Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol. 1997, 113, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Bird, C.R.; Smith, C.J.S.; Ray, J.A.; Moureau, P.; Bevan, M.W.; Bird, A.S.; Hughes, S.; Morris, P.C.; Grierson, D.; Schuch, W. The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Mol. Biol. 1988, 11, 651–662. [Google Scholar] [CrossRef] [PubMed]
- DellaPenna, D.; Bennett, A.B. In vitro synthesis and processing of tomato fruit polygalacturonase. Plant Physiol. 1988, 86, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Deytieux-Belleau, C.; Vallet, A.; Doneche, B.; Geny, L. Pectin methylesterase and polygalacturonase in the developing grape skin. Plant Physiol. Biochem. 2008, 46, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, Y.; Zhang, Z.; Ji, X.; Zhang, R.; Liu, D.; Gao, L.; Zhang, J.; Wang, B.; Wu, Y.; et al. Hypersensitive ethylene signaling and zmdpg1 expression lead to fruit softening and dehiscence. PLoS ONE 2013, 8, e58745. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cao, S.; Fang, X.; Mu, H.; Yang, H.; Wang, X.; Xu, Q.; Gao, H. Changes in fruit firmness, cell wall composition and cell wall degrading enzymes in postharvest blueberries during storage. Sci. Hortic. 2015, 188, 44–48. [Google Scholar] [CrossRef]
- Wei, J.; Ma, F.; Shi, S.; Qi, X.; Zhu, X.; Yuan, J. Changes and postharvest regulationof activity and gene expression of enzymes related to cell wall degradation inripening apple fruit. Postharvest Biol. Technol. 2010, 56, 147–154. [Google Scholar] [CrossRef]
- Pan, L.; Zeng, W.; Niu, L.; Lu, Z.; Liu, H.; Cui, G.; Zhu, Y.; Chu, J.; Li, W.; Fang, W.; et al. PpYUC11, a strong candidate gene for the stony hard phenotype in peach (Prunus persica L. Batsch), participates in IAA biosynthesis during fruit ripening. J. Exp. Bot. 2015, 66, 7031–7044. [Google Scholar] [CrossRef] [PubMed]
- Bapat, V.A.; Trivedi, P.K.; Ghosh, A.; Sane, V.A.; Ganapathi, T.R.; Nath, P. Ripening of fleshy fruit: Molecular insight and the role of ethylene. Biotechnol. Adv. 2010, 28, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Hayama, H.; Shimada, T.; Fujii, H.; Ito, A.; Kashimura, Y. Ethylene-regulation of fruit softening and softening-related genes in peach. J. Exp. Bot. 2006, 57, 4071–4077. [Google Scholar] [CrossRef] [PubMed]
- Hayama, H.; Tatsuki, M.; Ito, A.; Kashimura, Y. Ethylene and fruit softening in the stony hard mutation in peach. Postharvest Biol. Technol. 2006, 41, 16–21. [Google Scholar] [CrossRef]
- Layne, D.R.; Bassi, D. The Peach: Botany, Production and Uses; CABI: Oxfordshire, UK, 2008. [Google Scholar]
- Pfam. Avaliable online: http://pfam.Sanger.Ac.Uk/ (accessed on 9 September 2016).
- Database, P.G. Available online: https://www.Rosaceae.Org/species/prunus_persica/genome_v2.0.A1 (accessed on 9 September 2016).
- SMART. Available online: http://smart.Embl-heidelberg.De/ (accessed on 9 September 2016).
- Torki, M.; Mandaron, P.; Mache, R.; Falconet, D. Characterization of a ubiquitous expressed gene family encoding polygalacturonase in arabidopsis thaliana. Gene 2000, 242, 427–436. [Google Scholar] [CrossRef]
- GSDS. Avaliable online: http://gsds.Cbi.Pku.Edu.Cn/ (accessed on 9 September 2016).
- WebLogo. Avaliable online: http://weblogo.Berkeley.Edu/logo.Cgi (accessed on 9 September 2016).
- V3.0, S. Avaliable online: http://www.Cbs.Dtu.Dk/services/signalp (accessed on 9 September 2016).
- ExPASY. Avaliable online: http://www.Expasy.Org/tool.Html (accessed on 9 September 2016).
- MEME. Avaliable online: http://meme.Nbcr.Net/meme/cgi-bin/meme.Cgi (accessed on 9 September 2016).
- NCBI. Avaliable online: http://www.Ncbi.Nlm.Nih.Gov/ (accessed on 9 September 2016).
- Liu, R.H.; Meng, J.L. Mapdraw: A microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan 2003, 25, 317–321. [Google Scholar] [PubMed]
- Xing, L.; Zhang, D.; Li, Y.; Shen, Y.; Zhao, C.; Ma, J.; An, N.; Han, M. Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in apple (malus domestica borkh.). Plant Cell Physiol. 2015, 56, 2052–2068. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liguori, G.; Weksler, A.; Zutahi, Y.; Lurie, S.; Kosto, I. Effect of 1-methylcyclopropene on ripening of melting flesh peaches and nectarines. Postharvest Biol. Technol. 2004, 31, 263–268. [Google Scholar] [CrossRef]
- Cao, L.; Zhao, C.; Su, S.; Luo, C.; Han, M. The role of β-hexosaminidase in peach (prunus persica) fruit softening. Sci. Hortic. 2014, 169, 226–233. [Google Scholar] [CrossRef]




| Gene Name | Gene ID | Description | Deduced Polypeptide | Signal Peptide | Domains | ||
|---|---|---|---|---|---|---|---|
| Length (aa) | MW (kDa) | PI | |||||
| PpPG1 | ppa018113m | Polygalacturonase At1g48100 | 510 | 54.48 | 6.7 | + | I, II, III, IV |
| PpPG2 | ppa005391m | Polygalacturonase At1g48100 | 463 | 50.16 | 4.88 | − | I, II, III, IV |
| PpPG3 | ppa026115m | Exopolygalacturonase clone GBGE184 | 376 | 40.07 | 8.84 | − | I, II, III, IV |
| PpPG4 | ppa015208m | Exopolygalacturonase clone GBGE184 | 376 | 40.06 | 8.74 | − | I, II, III, IV |
| PpPG5 | ppa1027214m | - | 470 | 51.15 | 9.53 | + | I, II, III, IV |
| PpPG6 | ppa004002m | Polygalacturonase At1g48100 | 536 | 57.83 | 8.8 | + | I, II, III, IV |
| PpPG7 | ppa015822m | Probable polygalacturonase At3g15720 | 379 | 40.23 | 7.23 | − | I, II, III, IV |
| PpPG8 | ppa015678m | Probable polygalacturonase At3g15720 | 390 | 41.35 | 6.35 | + | I, II, III, IV |
| PpPG9 | ppa017135m | Polygalacturonase | 292 | 30.67 | 6.28 | − | I, II, III |
| PpPG10 | ppa026655m | Polygalacturonase | 375 | 40.83 | 9.09 | − | I, II, III, IV |
| PpPG11 | ppa018224m | Probable polygalacturonase | 463 | 50.68 | 6.56 | + | I, II, IV |
| PpPG12 | ppa020086m | Probable polygalacturonase | 517 | 55.99 | 8.91 | + | I, II, IV |
| PpPG13 | ppa005310m | Probable polygalacturonase | 467 | 51.58 | 5.74 | + | I, II, IV |
| PpPG14 | ppa014982m | Probable polygalacturonase At1g80170 | 449 | 48.85 | 8.74 | + | I, II, III, IV |
| PpPG15 | ppa018901m | Probable polygalacturonase | 391 | 42.72 | 6.66 | − | I, II, IV |
| PpPG16 | ppa005185m | Probable polygalacturonase | 473 | 52.28 | 8.73 | + | I, II, IV |
| PpPG17 | ppa024649m | Polygalacturonase At1g48100 | 437 | 47.60 | 5.95 | − | I, II, III, IV |
| PpPG18 | ppa021095m | Probable polygalacturonase At3g15720 | 621 | 65.91 | 6 | + | I, II, III, IV |
| PpPG19 | ppa021953m | Polygalacturonase | 376 | 39.46 | 8.35 | − | I, II, III, IV |
| PpPG20 | ppa025787m | Polygalacturonase | 392 | 40.98 | 9.28 | + | I, II, III, IV |
| PpPG21 | ppa006839m | Polygalacturonase | 393 | 41.33 | 6.24 | + | I, II, III, IV |
| PpPG22 | ppa006857m | Polygalacturonase | 393 | 41.26 | 6.24 | + | I, II, III, IV |
| PpPG23 | ppa005015m | Probable polygalacturonase | 481 | 52.39 | 6.36 | + | I, II, IV |
| PpPG24 | ppa004996m | Probable polygalacturonase | 482 | 52.45 | 5.81 | + | I, II, IV |
| PpPG25 | ppa005818m | Probable polygalacturonase | 442 | 48.32 | 7.12 | + | I, II, IV |
| PpPG26 | ppa019727m | Probable polygalacturonase At3g15720 | 382 | 41.13 | 8.66 | − | I, II, III, IV |
| PpPG27 | ppa025098m | Polygalacturonase | 384 | 40.72 | 7.54 | − | I, II, III, IV |
| PpPG28 | ppa014719m | Polygalacturonase At1g48100 | 419 | 45.94 | 8.27 | − | I, II, III, IV |
| PpPG29 | ppa025464m | Polygalacturonase | 381 | 40.56 | 8.76 | − | I, II, III, IV |
| PpPG30 | ppa016722m | Polygalacturonase | 432 | 46.30 | 8.72 | − | I, II, III, IV |
| PpPG31 | ppa018682m | Polygalacturonase | 371 | 39.42 | 8.17 | − | I, II, III, IV |
| PpPG32 | ppa018149m | Polygalacturonase | 307 | 33.04 | 8.18 | − | I, II, III |
| PpPG33 | ppa018308m | Polygalacturonase | 376 | 39.92 | 7.89 | − | I, II, III, IV |
| PpPG34 | ppa023489m | Probable polygalacturonase At3g15720 | 436 | 46.94 | 7.84 | + | I, II, III, IV |
| PpPG35 | ppa022427m | Polygalacturonase | 431 | 46.44 | 5.81 | − | I, II, III, IV |
| PpPG36 | ppa018519m | Probable polygalacturonase At3g15720 | 405 | 43.79 | 5.82 | + | I, II, III, IV |
| PpPG37 | ppa005960m | Probable polygalacturonase | 435 | 47.64 | 5.75 | + | I, II |
| PpPG38 | ppa004793m | Probable polygalacturonase | 491 | 55.03 | 9.15 | − | I, II, IV |
| PpPG39 | ppa021427m | Probable polygalacturonase At3g15720 | 383 | 41.32 | 7.92 | − | I, II, III, IV |
| PpPG40 | ppa007271m | Polygalacturonase | 375 | 38.92 | 8.51 | − | I, II, III, IV |
| PpPG41 | ppa020072m | Polygalacturonase | 395 | 42.03 | 9.52 | + | I, II, III, IV |
| PpPG42 | ppa016502m | Polygalacturonase | 403 | 42.82 | 9.24 | + | I, II, III, IV |
| PpPG43 | ppa019563m | Exopolygalacturonase | 387 | 41.55 | 8.93 | − | I, II, III, IV |
| PpPG44 | ppb019654m | Polygalacturonase | 382 | 30.80 | 8.81 | + | I, IV |
| PpPG45 | ppa015568m | Exopolygalacturonase clone GBGE184 | 482 | 50.57 | 6.32 | + | I, II, III, IV |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, M.; Zhang, Y.; Yan, X.; Han, M.; Li, J.; Li, F.; Li, F.; Zhang, D.; Zhao, C. Identification and Expression Analysis of Polygalacturonase Family Members during Peach Fruit Softening. Int. J. Mol. Sci. 2016, 17, 1933. https://doi.org/10.3390/ijms17111933
Qian M, Zhang Y, Yan X, Han M, Li J, Li F, Li F, Zhang D, Zhao C. Identification and Expression Analysis of Polygalacturonase Family Members during Peach Fruit Softening. International Journal of Molecular Sciences. 2016; 17(11):1933. https://doi.org/10.3390/ijms17111933
Chicago/Turabian StyleQian, Ming, Yike Zhang, Xiangyan Yan, Mingyu Han, Jinjin Li, Fang Li, Furui Li, Dong Zhang, and Caiping Zhao. 2016. "Identification and Expression Analysis of Polygalacturonase Family Members during Peach Fruit Softening" International Journal of Molecular Sciences 17, no. 11: 1933. https://doi.org/10.3390/ijms17111933






