Directed Evolution of Dunaliella salina Ds-26-16 and Salt-Tolerant Response in Escherichia coli
Abstract
:1. Introduction
2. Results
2.1. Construction of Random Mutant Library and Screening for Salt-Tolerance Variants
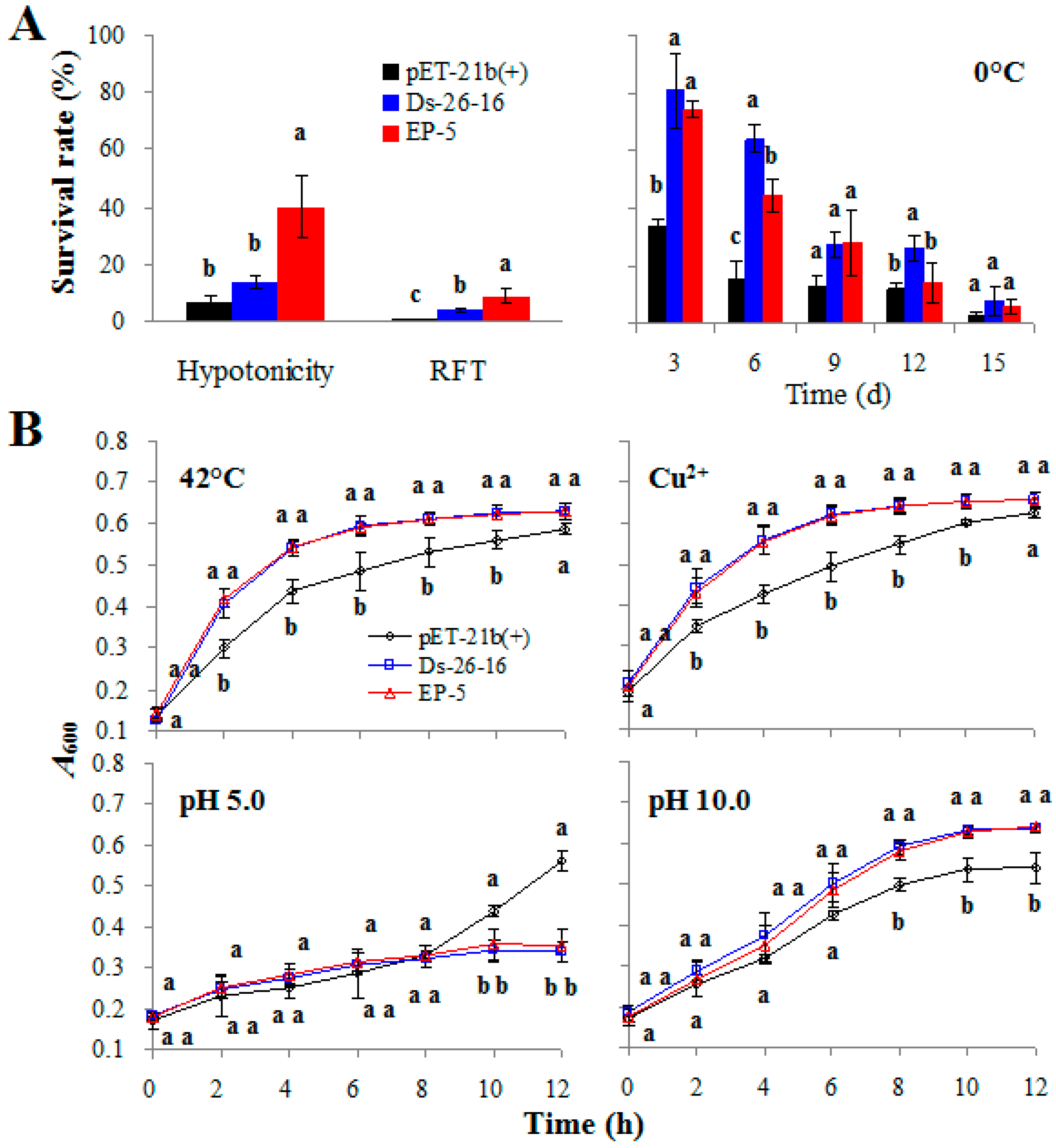
2.2. EP-5 Significantly Improved the Salt Tolerance to E. coli Transformant
2.3. EP-5 Conferred Other Abiotic Tolerances to E. coli Transformants
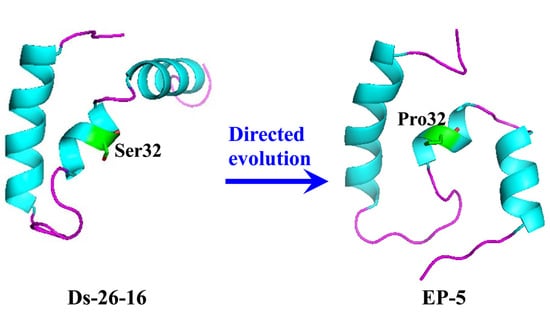
2.4. S32P Mutation Changed the Relationship of Two α-Helixes on N- and C-Terminal in EP-5
2.5. EP-5 Maintained the Normal Cell Morphology of E. coli Transformant
2.6. EP-5 Improved the Antioxidant Capacity of E. coli Transformant
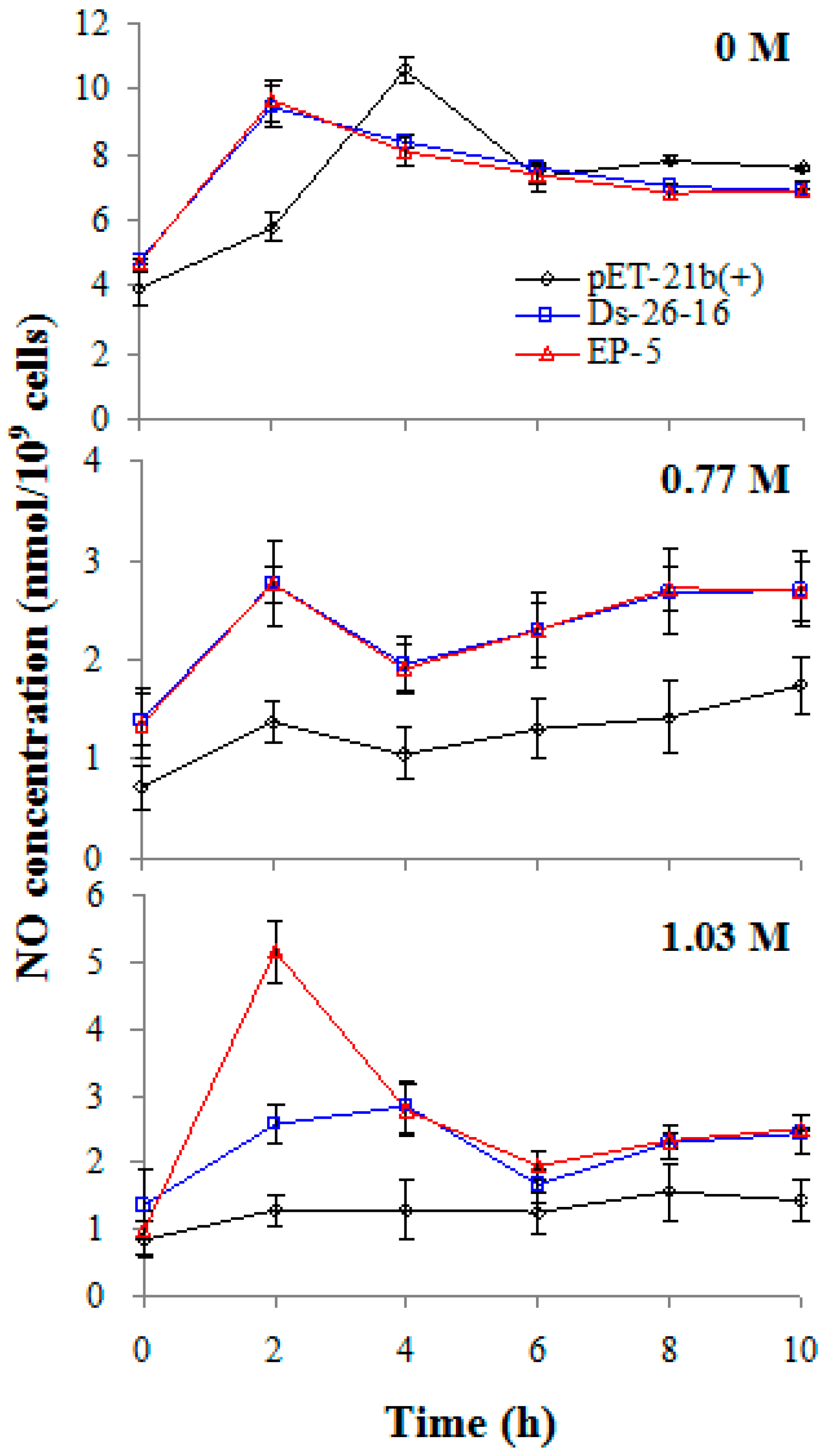
2.7. EP-5 Stimulated NO Synthesis in E. coli Transformant
3. Discussion
4. Materials and Methods
4.1. Plasmid, Chemical Reagents and Strains
4.2. Construction of Ds-26-16 Random Mutation Library
4.3. Screening for Ds-26-16 Variant
4.4. Stress Tolerance Analysis
4.5. Homology Modeling
4.6. Cell Surface Scanning with SEM
4.7. Antioxidant Enzyme Activity, MDA and NO Content Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Y.; Yuan, J.; Qin, H.; Zhang, W.G. Improvement of l-lactic acid production by osmotic-tolerant mutant of Lactobacillus casei at high temperature. Appl. Microbiol. Biotechnol. 2011, 89, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Lindemose, S.; O’Shea, C.; Jensen, M.K.; Skriver, K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef] [PubMed]
- Broun, P. Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 2004, 7, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Shukla, P.S.; Gupta, K.; Jha, B. Bioengineering for salinity tolerance in plants: State of the art. Mol. Biotechnol. 2013, 54, 102–123. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Meyer, M.M.; Meinhold, P.; Otey, C.R.; MacMillan, D.; Arnold, F.H. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 2005, 15, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Mangano, S.; Silberstein, S.; Santa-Maria, G.E. Point mutations in the barley HvHAK1 potassium transporter lead to improved K+-nutrition and enhanced resistance to salt stress. FEBS Lett. 2008, 582, 3922–3928. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Wang, G.; Ji, J.; Li, Y.; Gao, H.; Wu, J.; Guan, W. A chimeric vacuolar Na+/H+ antiporter gene evolved by DNA family shuffling confers increased salt tolerance in yeast. J. Biotechnol. 2015, 203, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Sung, B.H.; Yu, B.J.; Lee, J.H.; Lee, S.H.; Kim, M.S.; Koob, M.D.; Kim, S.C. Phenotypic engineering by reprogramming gene transcription using novel artificial transcription factors in Escherichia coli. Nucleic Acids Res. 2008, 36, e102. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Tyo, K.; Alper, H.; Klein-Marcuschamer, D.; Stephanopoulos, G. A high-throughput screen for hyaluronic acid accumulation in recombinant Escherichia coli transformed by libraries of engineered sigma factors. Biotechnol. Bioeng. 2008, 101, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chong, H.; Ching, C.B.; Jiang, R. Random mutagenesis of global transcription factor cAMP receptor protein for improved osmotolerance. Biotechnol. Bioeng. 2012, 109, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Alper, H.; Moxley, J.; Nevoigt, E.; Fink, G.R.; Stephanopoulos, G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 2006, 314, 1565–1568. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Bae, J.Y.; Lee, Y.M.; Kwon, H.; Moon, H.Y.; Kang, H.A.; Yee, S.B.; Kim, W.; Choi, W. Construction of Saccharomyces cerevisiae strains with enhanced ethanol tolerance by mutagenesis of the TATA-binding protein gene and identification of novel genes associated with ethanol tolerance. Biotechnol. Bioeng. 2011, 108, 1776–1787. [Google Scholar] [CrossRef] [PubMed]
- Kagaya, Y.; Hobo, T.; Murata, M.; Ban, A.; Hattori, T. Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 2002, 14, 3177–3189. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcriptional activator of novel ABRE dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Bazakos, C.; Manioudaki, M.E.; Therios, I.; Voyiatzis, D.; Kafetzopoulos, D.; Awada, T.; Kalaitzis, P. Comparative transcriptome analysis of two olive cultivars in response to NaCl-stress. PLoS ONE 2012, 7, e42931. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Zhao, L.; Hu, B.; Chen, X.; Zhang, F.; Zhu, Z.; Chen, D. Identifying novel salt-tolerant genes from Dunaliella salina using a Haematococcus pluvialis expression system. Plant Cell Tissue Organ. Cult. 2014, 117, 113–124. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, B.; Du, S.; Gao, S.; Chen, X.; Chen, D. Proteomic analyses reveal the mechanism of Dunaliella salina Ds-26–16 gene enhancing salt tolerance in Escherichia coli. PLoS ONE 2016, 11, e0153640. [Google Scholar]
- Hussain, S.S.; Kayani, M.A.; Amjad, M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol. Prog. 2011, 27, 297–306. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.A.; McDowell, D.A.; Blair, I.S. The pattern of pleiomorphism in stressed Salmonella virchow populations is nutrient and growth phase dependent. Lett. Appl. Microbiol. 2007, 45, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Courtois, C.; Besson, A.; Dahan, J.; Bourque, S.; Dobrowolska, G.; Pugin, A.; Wendehenne, D. Nitric oxide signalling in plants: Interplays with Ca2+ and protein kinase. J. Exp. Bot. 2008, 59, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.S. Protein Engineering: Protein Library Design, Folding, Databases, Design of Vaccines; LAP LAMBERT Academic Publishing: Saarbrücken, Germany, 2012; pp. 666–671. [Google Scholar]
- Koshkin, A.A. Syntheses and base-pairing properties of locked nucleic acid nucleotides containing hypoxanthine, 2,6-diaminopurine, and 2-aminopurine nucleobases. J. Org. Chem. 2004, 69, 3711–3718. [Google Scholar] [CrossRef] [PubMed]
- Hobert, O. Gene regulation by transcription factors and microRNAs. Science 2008, 319, 1785–1786. [Google Scholar] [CrossRef] [PubMed]
- Boniolo, F.S.; Rodrigues, R.C.; Delatorre, E.O.; da Silveira, M.M.; Flores, V.M.; Berbert-Molina, M.A. Glycine betaine enhances growth of nitrogen-fixing bacteria Gluconacetobacter diazotrophicus PAL5 under saline stress conditions. Curr. Microbiol. 2009, 59, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Mattick, K.L.; Rowbury, R.J.; Humphrey, T.J. Morphological changes to Escherichia coli 0157:H7, commensal E. coli and Salmonella spp. in response to marginal growth conditions, with special reference to mildly stressing temperatures. Sci. Prog. 2003, 86, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wei, G.; Li, J.; Qian, Q.; Yu, J. Silicon alleviates salt stress and increases antioxidant enzymes activity in leaves of salt-stressed cucumber (Cucumis sativus L.). Plant Sci. 2004, 167, 527–533. [Google Scholar] [CrossRef]
- David, A.; Yadav, S.; Baluska, F.; Bhatla, S.C. Nitric oxide accumulation and protein tyrosine nitration as a rapid and long distance signaling response to salt stress in sunflower seedlings. Nitric Oxide 2015, 50, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.G.; Tian, Q.Y.; Zhang, W.H. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 2007, 144, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Chen, Q.; Liu, Q.; Zhang, W.; Ding, R. Exogenous silicon (Si) increases antioxidant enzyme activity and reduces lipid peroxidation in roots of salt-stressed barley (Hordeum vulgare L.). J. Plant Physiol. 2003, 160, 1157–1164. [Google Scholar] [CrossRef] [PubMed]



| NaCl (M) | pET-21b(+) | Ds-16-16 | EP-5 |
|---|---|---|---|
| 0 | 2.23 ± 0.61 a | 2.37 ± 0.74 a | 2.32 ± 0.86 a |
| 0.51 | 31.25 ± 9.84 a | 2.70 ± 0.87 b | 2.36 ± 0.72 b |
| 1.03 | 7.29 ± 6.83 a | 4.20 ± 1.72 b | 4.77 ± 2.95 b |
| 1.54 | 1.81 ± 1.22 c | 3.06 ± 1.78 b | 3.74 ± 2.31 a |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Dong, Y.; Hong, X.; Pang, X.; Chen, D.; Chen, X. Directed Evolution of Dunaliella salina Ds-26-16 and Salt-Tolerant Response in Escherichia coli. Int. J. Mol. Sci. 2016, 17, 1813. https://doi.org/10.3390/ijms17111813
Guo Y, Dong Y, Hong X, Pang X, Chen D, Chen X. Directed Evolution of Dunaliella salina Ds-26-16 and Salt-Tolerant Response in Escherichia coli. International Journal of Molecular Sciences. 2016; 17(11):1813. https://doi.org/10.3390/ijms17111813
Chicago/Turabian StyleGuo, Yuan, Yanping Dong, Xiao Hong, Xiaonan Pang, Defu Chen, and Xiwen Chen. 2016. "Directed Evolution of Dunaliella salina Ds-26-16 and Salt-Tolerant Response in Escherichia coli" International Journal of Molecular Sciences 17, no. 11: 1813. https://doi.org/10.3390/ijms17111813