Study on Dicyandiamide-Imprinted Polymers with Computer-Aided Design
Abstract
:1. Introduction
2. Results and Discussion
2.1. Selection of the Appropriate Calculated Method
2.2. Forecast of the Active Sites for the Template and Function Monomers
2.3. Selection of Functional Monomer Based on Theoretical Calculations
2.4. Selection of Cross-Linker Based on Theoretical Calculations
2.5. Adsorption Property of the MIPs
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Computational Methods
3.3. Preparation of MIPs and NIPs
3.4. Analysis of Adsorption Property
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MIPs | Molecularly Imprinted Polymers |
| DCD | Dicyandiamide |
| MAA | Methacrylic Acid |
| PETA | Pentaerythritol Triacrylater |
| AM | Acrylamide |
| MBA | N,N’-methylenebisacrylamide |
| IA | Itaconic Acid |
| DVB | Divinylbenzene |
| EGDMA | Ethylene Glycol Dimethacrylate |
| TRIM | Trimethylolpropane Trimethylacrylate |
| MEPs | Molecular Electrostatic Potentials |
| NIPs | Non-imprinted Polymers |
| SEM | Scanning Electron Microscope |
| MAM | Melamine |
| CYR | Cyromazine |
| TRI | Trithiocyanuric |
| BSSE | Basis Set Superposition Error |
References
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef] [PubMed]
- Bedwell, T.; Whitcombe, M. Analytical application of MIPs in diagnostic assay: Future perspectives. Anal. Bioanal. Chem. 2016, 408, 1735–1751. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, T.; Cui, L.; Jide, W.; Piletska, E.V.; Guerreiro, A.R.; Piletsky, S.A. Rational design and synthesis of water-compatible molecularly imprinted polymers for selective solid phase extraction of amiodarone. Anal. Chim. Acta 2012, 709, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Nestić, M.; Babić, S.; Pavlović, D.M.; Sutlović, D. Molecularly imprinted solid phase extraction for simultaneous determination of ∆9-tetrahydrocannabinol and its main metabolites by gas chromatography–mass spectrometry in urine samples. Forensic Sci. Int. 2013, 231, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.L.; Wang, X.C.; Lin, X.C.; Wu, X.P.; Xie, Z.H. A molecularly imprinted monolith for the fast chira separation of antiparasitic drugs by pressurized CEC. J. Sep. Sci. 2010, 33, 2123–2130. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Bondi, M.C.; Benito-Peña, M.E.; Urraca, J.L.; Orellana, G. Immuno-like assays and biomimetic microchips. Top. Curr. Chem. 2012, 325, 111–164. [Google Scholar] [PubMed]
- Chianella, I.; Guerreiro, A.; Moczko, E.; Caygill, J.S.; Piletska, E.V.; Sansalvador, I.M.P.D.V.; Whitcombe, M.J.; Piletsky, S.A. Direct replacement of antibodies with molecularly imprinted polymer nanoparticles in ELISA-development of a novel assay for vancomycin. Anal. Chem. 2013, 85, 8462–8468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, R.N.; Kou, L.J.; Chen, Z.P.; Qin, W. Molecularly imprinted nanoparticles based potentiometric sensor with a nanomolar detection limit. Sens. Actuators B-Chem. 2013, 188, 972–977. [Google Scholar] [CrossRef]
- Granado, V.L.V.; Gutiérrez-Capitán, M.; Fernández-Sánchez, C.; Gomes, M.T.S.R.; Rudnitskaya, A.; Jimenez-Jorquer, C. Thin-film electrochemical sensor for diphenylamine detection using molecularly imprinted polymers. Anal. Chim. Acta 2014, 809, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Kupai, J.; Rojik, E.; Huszthy, P.; Szekely, G. Role of chirality and macroring in imprinted polymers with enantiodiscriminative power. ACS Appl. Mater. Interfaces 2015, 7, 9516–9525. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, X.W.; Chen, L.X.; Zhang, Y.K. Preparation of core-shell magnetic molecularly imprinted polymer nanoparticles for recognition of bovine hemoglobin. Chem. Asian J. 2009, 4, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Razali, M.; Kim, J.F.; Attfield, M.; Budd, P.M.; Drioli, E.; Lee, Y.M.; Szekely, G. Sustainable wastewater treatment and recycling in membrane manufacturing. Green Chem. 2015, 17, 5196–5205. [Google Scholar] [CrossRef]
- Del Sole, R.; Lazzoi, M.R.; Arnone, M.; Della Sala, F.; Cannoletta, D.; Vasapollo, G. Experimental and computational studies on non-covalent imprinted microspheres as recognition system for nicotinamide. Molecules 2009, 14, 2632–2649. [Google Scholar] [CrossRef] [PubMed]
- Pietrzyk, A.; Kutner, W.; Chitta, R.; Zandler, M.E.; D’Souza, F.; Sannicolò, F.; Mussini, P.R. Melamine acoustic chemosensor based on molecularly imprinted polymer film. Anal. Chem. 2009, 81, 10061–10070. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.K.; Cao, Y.; Ma, P.F.; Qiu, C.X.; Xu, W.Z.; Liu, H.; Huang, W.H. Rational design and preparation of magnetic imprinted polymers for removal of indole by molecular simulation and improved atom transfer radical polymerization. RSC Adv. 2014, 4, 605–616. [Google Scholar] [CrossRef]
- Wei, S.; Jakusch, M.; Mizaikoff, B. Investigating the mechanisms of 17β-estradiol imprinting by computational prediction and spectroscopic analysis. Anal. Bioanal. Chem. 2007, 389, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Yañez, F.; Chianella, I.; Piletsky, S.A.; Concheiro, A.; Alvarez-Lorenzo, C. Computational modeling and molecular imprinting for the development of acrylic polymers with high affinity for bile salts. Anal. Chim. Acta 2010, 659, 178–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, H.; Morimoto, S.; Hayatsu, M.; Hayakawa, A.; Sudo, S.; Yagi, K. Nitrification, ammonia-oxidizing communities, and N2O and CH4 fluxes in an imperfectly drained agricultural field fertilized with coated urea with and without dicyandiamide. Biol. Fertil. Soils 2013, 49, 213–223. [Google Scholar] [CrossRef]
- Di, H.J.; Cameron, K.C. The use of a nitrification inhibitor, dicyandiamide (DCD), to decrease nitrate leaching and nitrous oxide emissions in a simulated grazed and irrigated grassland. Soil Use Manag. 2002, 18, 395–403. [Google Scholar] [CrossRef]
- Liu, H.L.; Zhou, K.W.; Wu, D.; Wang, J.; Sun, B.G. A novel quantum dots-labeled on the surface of molecularly imprinted polymer for turn-off optosensing of dicyandiamide in dairy products. Biosens. Bioelectron. 2016, 77, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Song, H.; Liu, J.; Liu, Y.; Li, L.; Tang, H.; Li, Y.C. Preparation of molecularly imprinted polymer with double templates for rapid simultaneous determination of melamine and dicyandiamide in dairy products. Talanta 2015, 134, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Khana, M.S.; Pala, S.; Krupadam, R.J. Computational strategies for understanding the nature of interaction in dioxin imprinted nanoporous trappers. J. Mol. Recognit. 2015, 28, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Diñeiro, Y.; Menéndez, M.I.; Blanco-López, M.C.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tunñón-Blanco, P. Computational approach to the rational design of molecularly imprinted polymers for voltammetric sensing of homovanillic acid. Anal. Chem. 2005, 77, 6741–6746. [Google Scholar] [CrossRef] [PubMed]
- Piacham, T.; Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Synthesis and computational investigation of molecularly imprinted nanospheres for selective recognition of alpha-tocopherol succinate. EXCLI J. 2013, 12, 701–718. [Google Scholar] [PubMed]
- Liu, J.B.; Shi, Y.; Sun, J.N.; Tang, S.S.; Hu, Y.H.; Jin, R.F. A theoretical study on the interaction between melamine and acrylamide functional monomer in molecularly imprinted polymers. Food Sci. 2013, 34, 96–101. [Google Scholar]
- Wang, Y.; Liu, J.B.; Tang, S.S.; Jin, R.F. Preparation of melamine molecularly imprinted polymer by computer-aided design. J. Sep. Sci. 2015, 38, 2647–2654. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.B.; Tang, S.S.; Dai, Z.Q.; Jin, R.F. Theoretical research on self-assembly system of molecularly imprinted polymers formed by melamine and trifluoromethacrylic acid. Struct. Chem. 2016, 27, 897–905. [Google Scholar] [CrossRef]
- Cheng, X.L.; Li, L.Q.; Zhao, Y.Y.; Wang, C.A. Absorption and emission spectroscopic characteristics of dipterex and its molecularly imprinted recognition: A TD-DFT investigation. Chem. Phys. Lett. 2016, 652, 93–97. [Google Scholar] [CrossRef]
- Huynh, T.P.; Sosnowska, M.; Sobczak, J.W.; Kc, C.B.; Nesterov, V.N.; D'Souza, F.; Kutner, W. Simultaneous chronoamperometry and piezoelectric microgravimetry determination of nitroaromatic explosives using molecularly imprinted thiophene polymers. Anal. Chem. 2013, 85, 8361–8368. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.B.; Shi, Y.; Tang, S.S.; Jin, R.F. Theoretical and experimental research on the self-assembled system of molecularly imprinted polymers formed by salbutamol and methacrylic acid. J. Sep. Sci. 2015, 38, 1065–1071. [Google Scholar]
- Hughes, E.W. The crystal structure of dicyandiamide. J. Am. Chem. Soc. 1940, 62, 1258–1267. [Google Scholar] [CrossRef]
- Pogány, P.; Razali, M.; Szekely, G. Experimental and theoretical investigation of the complexation of methacrylic acid and diisopropyl urea. Spectrochim. Acta Part A 2017, 170, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09 Suite of Programs; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Boys, S.F.; Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970, 19, 553–566. [Google Scholar] [CrossRef]
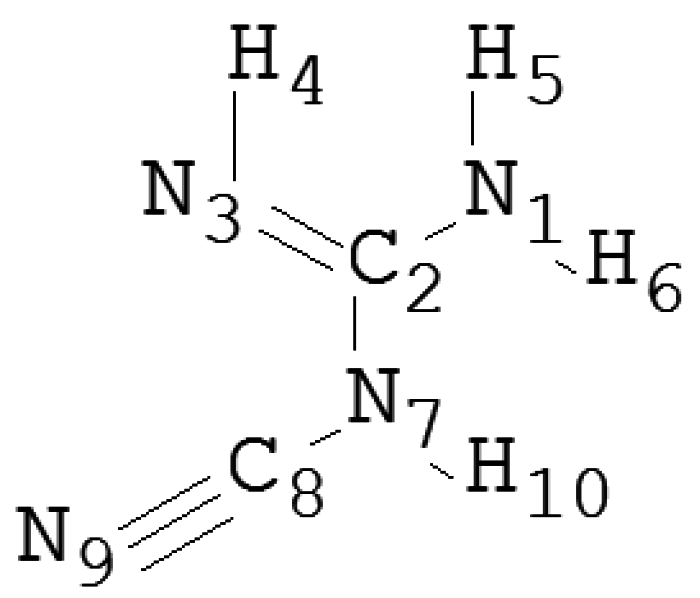
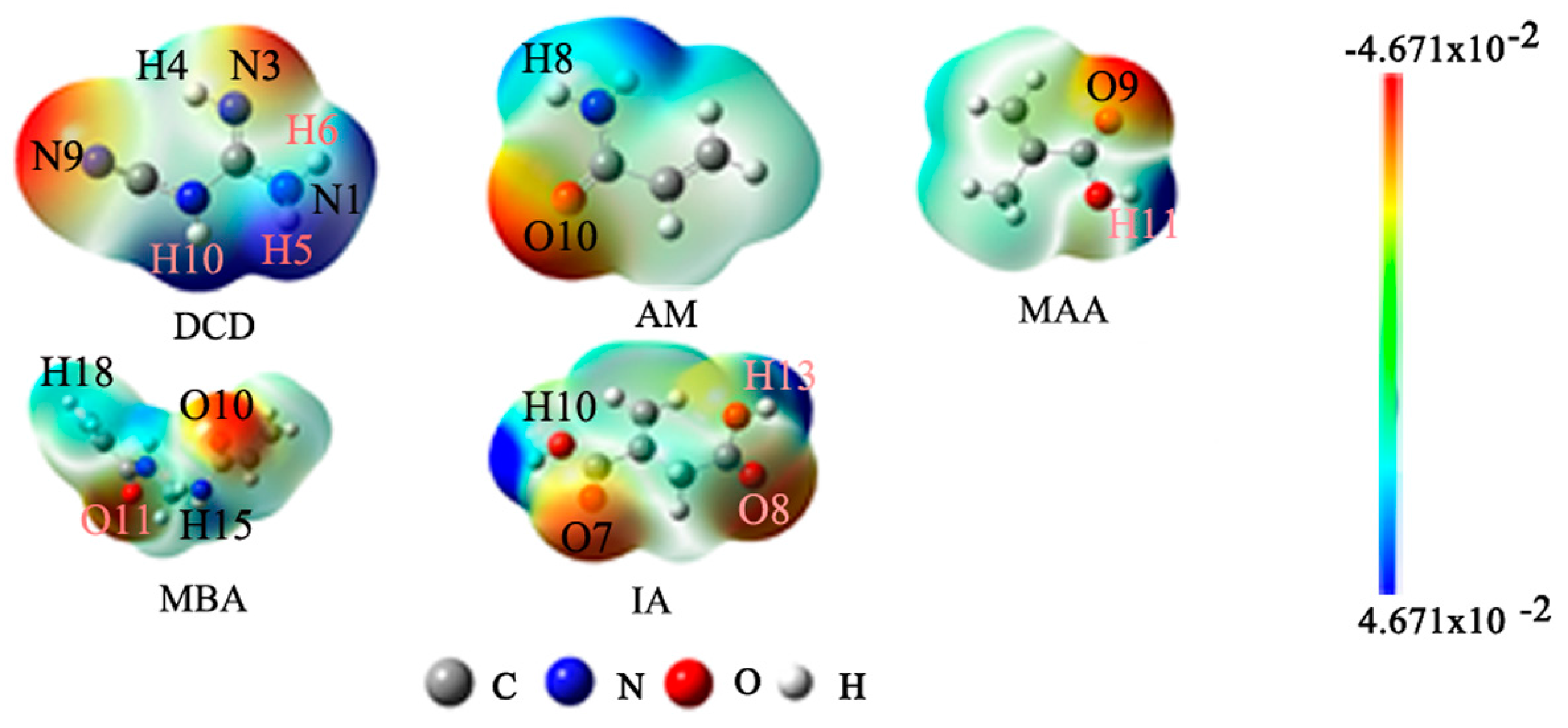
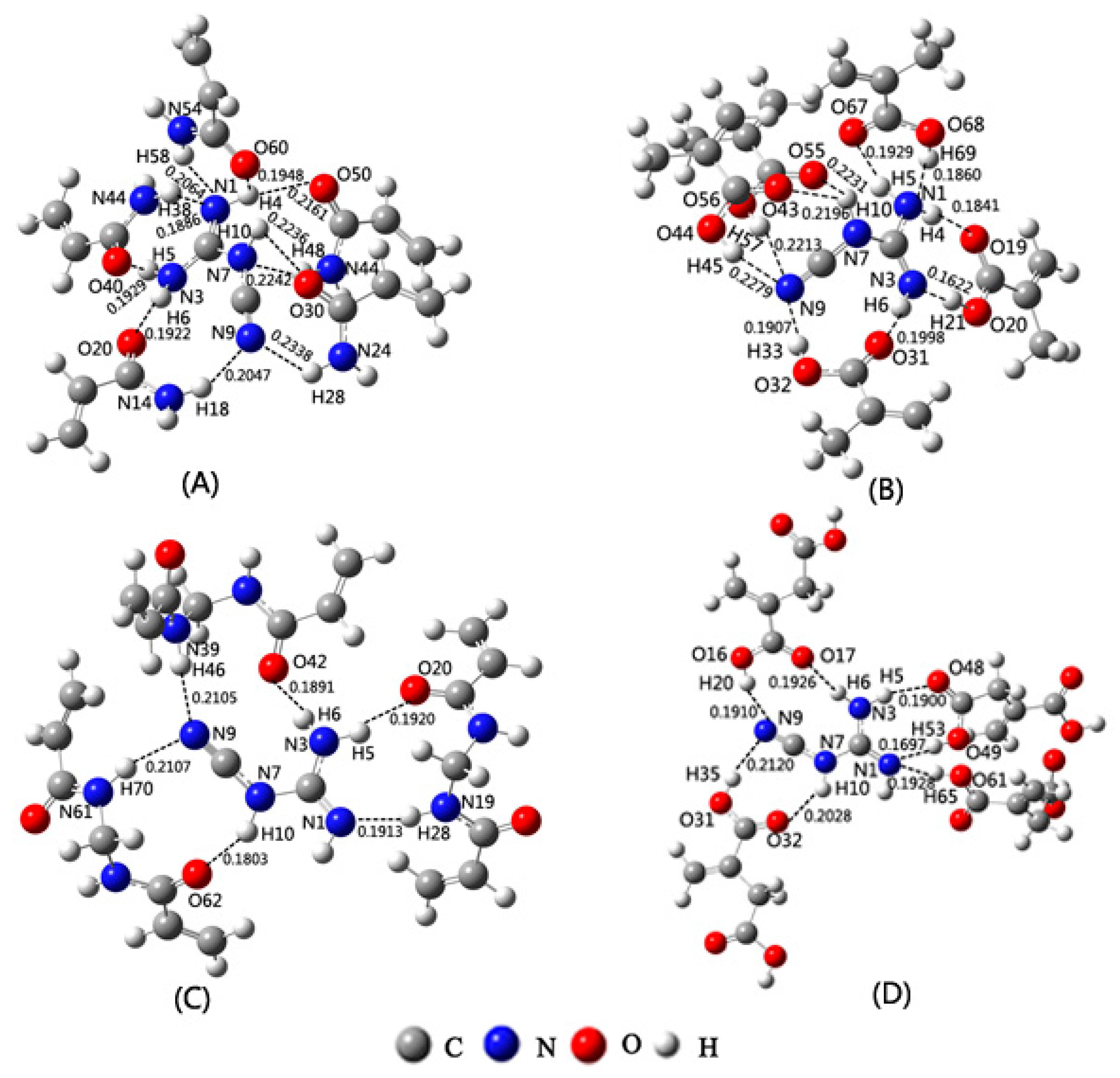


| Species | wB97XD | B3LYP | PBE1PBE | LC-wPBE | M062X | Experimental |
|---|---|---|---|---|---|---|
| R(Å) | ||||||
| N1–C2 | 1.27 | 1.27 | 1.27 | 1.27 | 1.28 | 1.34 |
| N3–C2 | 1.37 | 1.37 | 1.36 | 1.36 | 1.37 | 1.37 |
| N7–C2 | 1.42 | 1.42 | 1.42 | 1.42 | 1.42 | 1.36 |
| N7–C8 | 1.33 | 1.33 | 1.33 | 1.33 | 1.33 | 1.28 |
| N9–C8 | 1.16 | 1.17 | 1.16 | 1.16 | 1.18 | 1.22 |
| Φ(°) | ||||||
| N1–C2–N3 | 123 | 123 | 123 | 123 | 123 | 124 |
| N1–C2–N7 | 121 | 121 | 121 | 121 | 121 | 120 |
| N3–C2–N7 | 115 | 115 | 115 | 115 | 115 | 116 |
| C2–N7–C8 | 121 | 121 | 121 | 121 | 121 | 120 |
| RatioComplexes | DCD-AM | DCD-MAA | DCD-MBA | DCD-IA |
|---|---|---|---|---|
| 1:1 | 10.33 | 73.60 | 0.05 | 3.28 |
| 1:2 | −40.03 | −59.09 | −55.06 | −42.148 |
| 1:3 | −93.73 | −106.41 | −114.47 | −120.14 |
| 1:4 | −122.02 | −143.47 | / | −134.45 |
| 1:5 | −151.75 | −191.40 | / | / |
| Species | EGDMA | DVB | PETA | TRIM |
|---|---|---|---|---|
| DCD | −36.49 | −34.92 | −33.61 | −53.30 |
| MAA | −23.63 | −16.02 | −43.58 | −37.54 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, D.; Wang, Y.; Li, S.; Li, Y.; Zhang, M.; Li, Y.; Tian, W.; Liu, J.; Tang, S.; Li, B.; et al. Study on Dicyandiamide-Imprinted Polymers with Computer-Aided Design. Int. J. Mol. Sci. 2016, 17, 1750. https://doi.org/10.3390/ijms17111750
Liang D, Wang Y, Li S, Li Y, Zhang M, Li Y, Tian W, Liu J, Tang S, Li B, et al. Study on Dicyandiamide-Imprinted Polymers with Computer-Aided Design. International Journal of Molecular Sciences. 2016; 17(11):1750. https://doi.org/10.3390/ijms17111750
Chicago/Turabian StyleLiang, Dadong, Yan Wang, Songyang Li, Yuqing Li, Miliang Zhang, Yang Li, Weishuai Tian, Junbo Liu, Shanshan Tang, Bo Li, and et al. 2016. "Study on Dicyandiamide-Imprinted Polymers with Computer-Aided Design" International Journal of Molecular Sciences 17, no. 11: 1750. https://doi.org/10.3390/ijms17111750





