Impact of SNPs on Protein Phosphorylation Status in Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Results
2.1. SNPs in Rice Genome Ver. 4.0 and 7.0
2.2. nsSNPs in Rice Genome Ver. 7.0
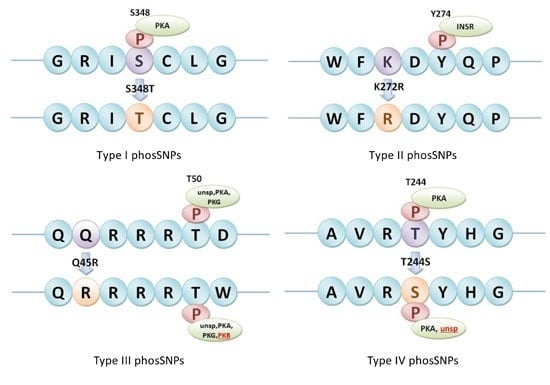
2.3. Prediction of PhosSNPs in Rice Genome
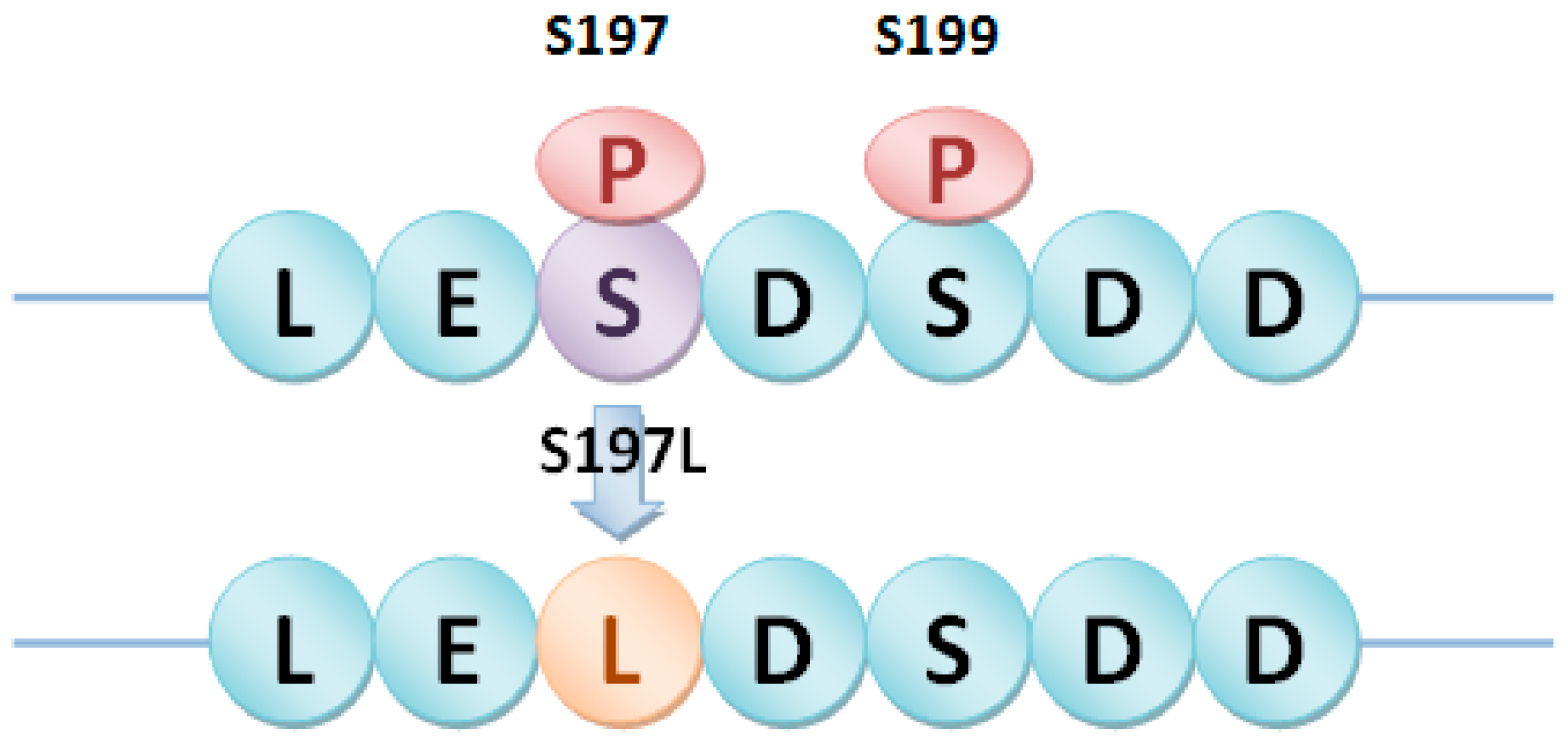
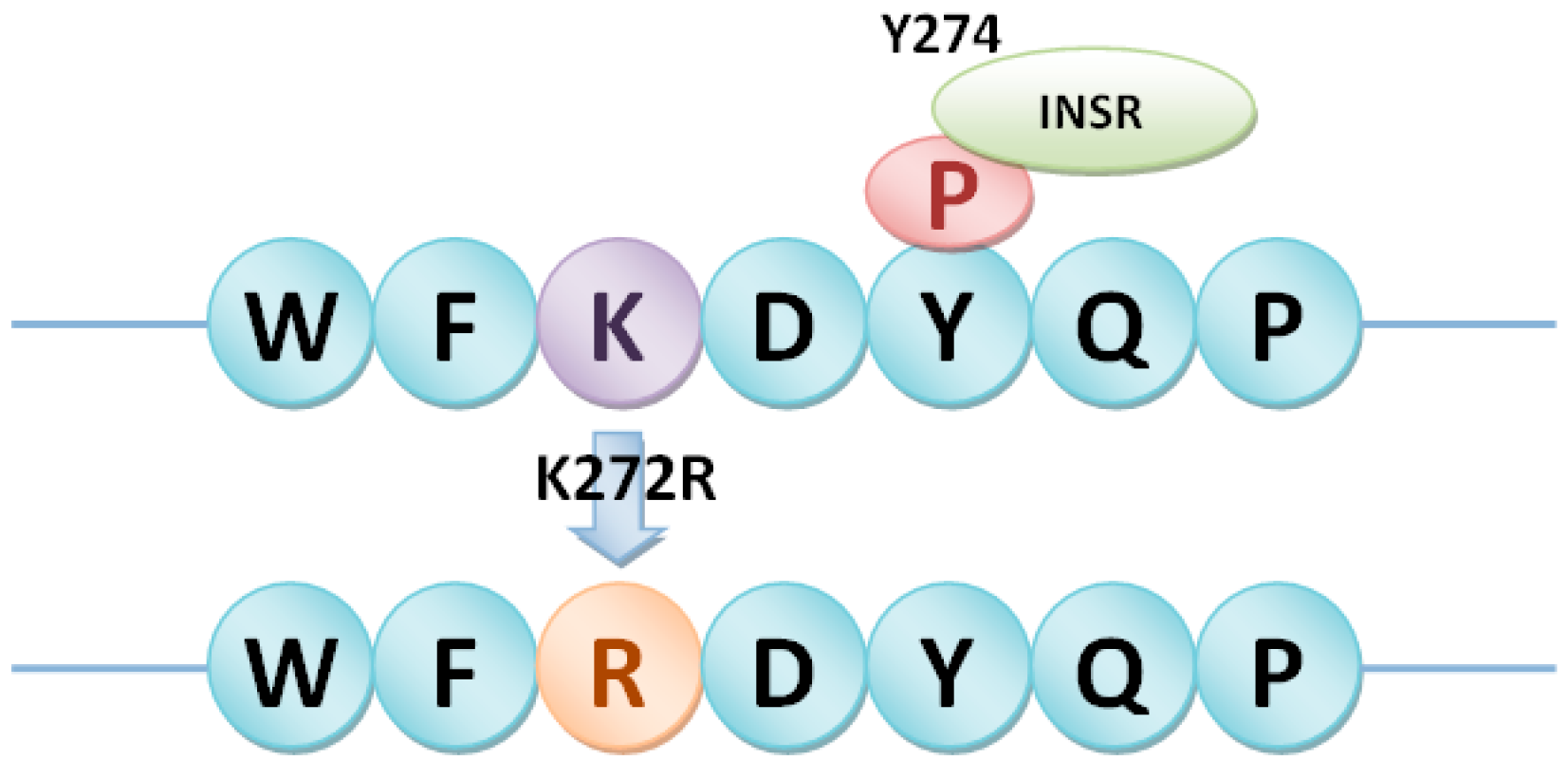
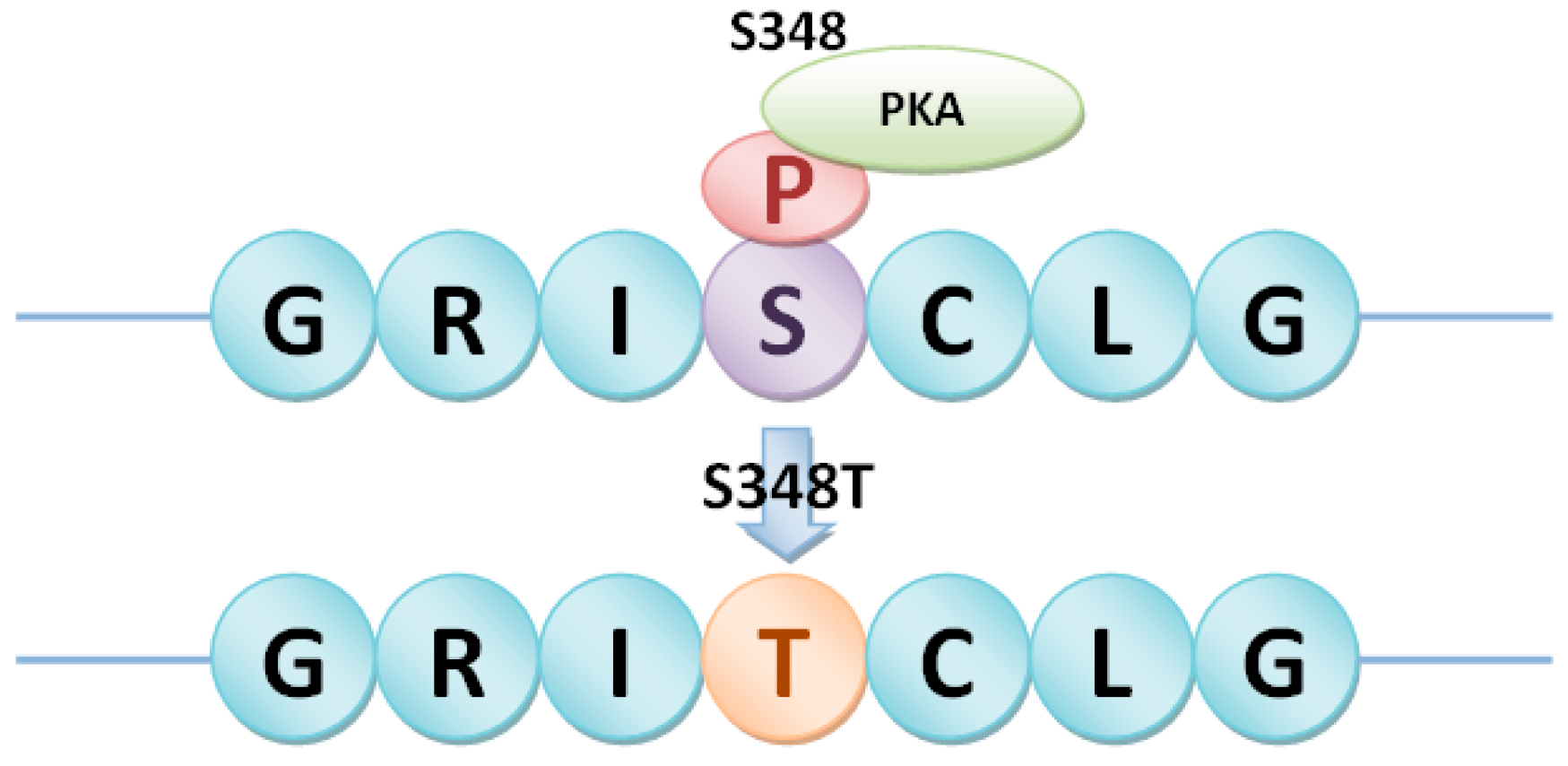
2.4. PhosSNPs in Heterotrimeric G Proteins
3. Discussion
4. Materials and Methods
4.1. SNPs in Rice Genome 4.0 and 7.0
4.2. Prediction of Phosphorylation Sites in Rice Proteome
4.3. Detection of Different Types of phosSNPs
4.4. PhosSNPs in Heterotrimeric G Proteins
4.5. Database Construction
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.A.; Ricke, D.; Lan, T.H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef] [PubMed]
- Yonemaru, J.; Choi, S.H.; Sakai, H.; Ando, T.; Shomura, A.; Yano, M.; Wu, J.; Fukuoka, S. Genome-wide indel markers shared by diverse Asian rice cultivars compared to Japanese rice cultivar 'Koshihikari'. Breed. Sci. 2015, 65, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Wang, J.; Zeigler, R.S. The 3000 rice genomes project: New opportunities and challenges for future rice research. Gigascience 2014, 3, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Nasu, S.; Suzuki, J.; Ohta, R.; Hasegawa, K.; Yui, R.; Kitazawa, N.; Monna, L.; Minobe, Y. Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryzarufipogon) and establishment of SNP markers. DNA Res. 2002, 9, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Brooks, L.D.; Chakravarti, A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998, 8, 1229–1231. [Google Scholar] [PubMed]
- Lyu, J.; Zhang, S.; Dong, Y.; He, W.; Zhang, J.; Deng, X.; Zhang, Y.; Li, X.; Li, B.; Huang, W.; et al. Analysis of elite variety tag SNPs reveals an important allele in upland rice. Nat. Commun. 2013, 4, 2138. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Gross, B.L.; Olsen, K.M. Genetic perspectives on crop domestication. Trends Plant Sci. 2010, 15, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Savas, S.; Ozcelik, H. Phosphorylation states of cell cycle and DNA repair proteins can be altered by the nsSNPs. BMC Cancer 2005, 5, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, G.M.; Song, P.; Kim, K.W.; Oh, K.S.; Park, K.J.; Kim, J.H. Genome-wide analysis to predict protein sequence variations that change phosphorylation sites or their corresponding kinases. Nucleic Acids Res. 2009, 37, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Erxleben, C.; Liao, Y.; Gentile, S.; Chin, D.; Gomez-Alegria, C.; Mori, Y.; Birnbaumer, L.; Armstrong, D.L. Cyclosporin and Timothy syndrome increase mode 2 gating of CaV1.2 calcium channels through aberrant phosphorylation of S6 helices. Proc. Natl. Acad. Sci. USA 2006, 103, 3932–3937. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Jiang, C.; Gao, X.; Liu, Z.; Yuan, Z.; Jin, C.; Wen, L.; Zhang, Z.; Xue, Y.; Yao, X. PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol. Cell. Proteom. 2010, 9, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Song, Q.; Tao, H.; Wang, W.; Wan, W.; Huang, J.; Xu, C.; Chebii, V.; Kitony, J.; Que, S.; et al. Rice_Phospho 1.0: A new rice-specific SVM predictor for protein phosphorylation sites. Sci. Rep. 2015, 5, 11940. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Li, J. Proteomic analysis of phosphoproteins regulated by abscisic acid in rice leaves. Biochem. Biophys. Res. Commun. 2008, 371, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Feltus, F.A.; Wan, J.; Schulze, S.R.; Estill, J.C.; Jiang, N.; Paterson, A.H. An SNP resource for rice genetics and breeding baseFd on subspecies indica and japonica genome alignments. Genome Res. 2004, 14, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Parida, S.K.; Mukerji, M.; Singh, A.K.; Singh, N.K.; Mohapatra, T. SNPs in stress-responsive rice genes: Validation, genotyping, functional relevance and population structure. BMC Genom. 2012, 13, 426. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Han, G.; He, H.; Li, J. Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem. Biophys. Res. Commun. 2009, 379, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Sicilia, M.N.; Trusov, Y.; Maruta, N.; Chakravorty, D.; Zhang, Y.; Botella, J.R. Heterotrimeric G proteins interact with defense-related receptor-like kinases in Arabidopsis. J. Plant Physiol. 2015, 188, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Trusov, Y.; Botella, J.R. Plant G-Proteins Come of Age: Breaking the Bond with Animal Models. Front. Chem. 2016, 4, 24. [Google Scholar] [CrossRef] [PubMed]
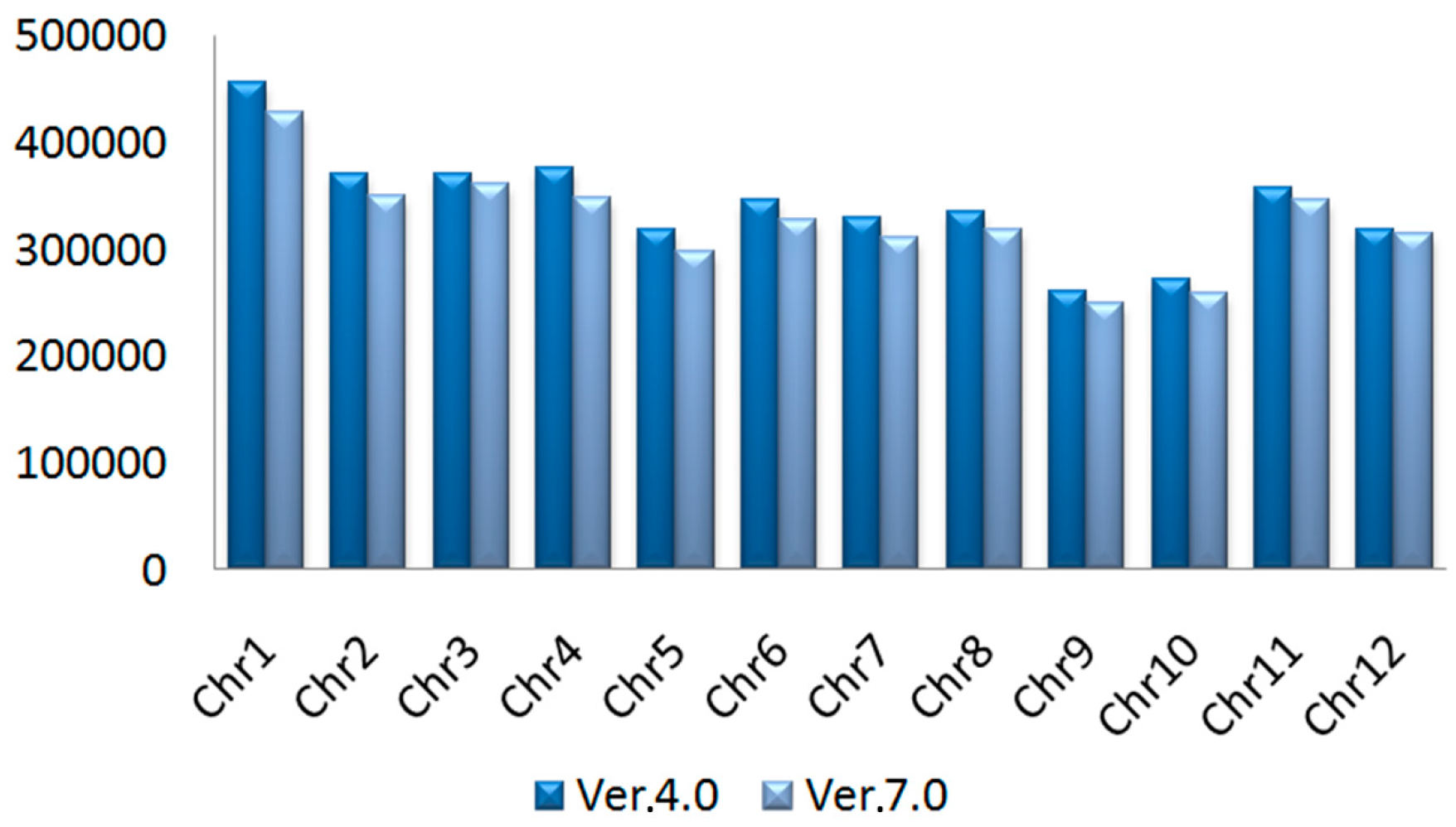


| Chromosome | Inter-Gene | Intron | UTR | sSNP | nsSNP | Number of Proteins Holding SNPs |
|---|---|---|---|---|---|---|
| Chr1 | 260,587 | 80,970 | 18,498 | 32,343 | 35,815 | 5709 |
| Chr2 | 212,543 | 65,908 | 14,728 | 24,573 | 32,379 | 4693 |
| Chr3 | 213,338 | 66,415 | 15,408 | 36,203 | 28,847 | 4771 |
| Chr4 | 207,656 | 61,928 | 11,237 | 31,795 | 34,520 | 4531 |
| Chr5 | 181,707 | 51,407 | 10,337 | 21,797 | 32,094 | 4034 |
| Chr6 | 202,075 | 58,375 | 10,506 | 22,845 | 33,679 | 4190 |
| Chr7 | 190,062 | 54,368 | 10,492 | 25,042 | 30,964 | 3884 |
| Chr8 | 194,303 | 56,258 | 10,264 | 22,845 | 33,969 | 3779 |
| Chr9 | 151,668 | 43,941 | 7336 | 18,703 | 26,741 | 3031 |
| Chr10 | 161,059 | 43,460 | 7593 | 22,062 | 25,383 | 3024 |
| Chr11 | 210,082 | 60,511 | 9046 | 31,228 | 34,399 | 3716 |
| Chr12 | 193,270 | 52,636 | 8547 | 24,792 | 35,775 | 3599 |
| Total | 2,378,350 | 696,177 | 133,992 | 314,228 | 384,565 | 48,961 |
| Chromosome | Type I (−) | Type I (+) | Type II (−) | Type III | Type IV |
|---|---|---|---|---|---|
| Chr1 | 2803 | 0 | 1411 | 7996 | 3215 |
| Chr2 | 2608 | 1 | 1374 | 6778 | 2693 |
| Chr3 | 2065 | 3 | 1034 | 6004 | 2491 |
| Chr4 | 2473 | 0 | 1284 | 6296 | 2513 |
| Chr5 | 2900 | 1 | 1466 | 6844 | 2878 |
| Chr6 | 3015 | 0 | 1348 | 7528 | 2852 |
| Chr7 | 2351 | 0 | 1028 | 6155 | 2455 |
| Chr8 | 3054 | 2 | 1491 | 7174 | 2949 |
| Chr9 | 2170 | 1 | 921 | 5299 | 2123 |
| Chr10 | 1805 | 2 | 910 | 4842 | 1916 |
| Chr11 | 2423 | 0 | 1021 | 6347 | 2630 |
| Chr12 | 3238 | 1 | 1327 | 7102 | 2991 |
| Total | 25,500 | 11 | 14,615 | 78,365 | 31,706 |
| Subunits | LOC ID | nsSNP ID | Nucleotide Mutation | Animo Acid Mutation |
|---|---|---|---|---|
| Gα | LOC_Os05g26890 | SNP050177186 | T/C | K272R |
| Gβ | LOC_Os03g46650 | SNP030274451 | A/G | S348T |
| SNP030274452 | T/C | R279G | ||
| SNP030274453 | T/A | T244S | ||
| SNP030274454 | A/G | N217T | ||
| Gγ2 | LOC_Os02g04520 | SNP020015843 | A/G | Q45R |
| SNP020015851 | G/T | R137L |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Chen, L.; Tao, H.; Huang, J.; Xu, C.; Li, L.; Ma, S.; Tian, T.; Liu, W.; Xue, L.; et al. Impact of SNPs on Protein Phosphorylation Status in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2016, 17, 1738. https://doi.org/10.3390/ijms17111738
Lin S, Chen L, Tao H, Huang J, Xu C, Li L, Ma S, Tian T, Liu W, Xue L, et al. Impact of SNPs on Protein Phosphorylation Status in Rice (Oryza sativa L.). International Journal of Molecular Sciences. 2016; 17(11):1738. https://doi.org/10.3390/ijms17111738
Chicago/Turabian StyleLin, Shoukai, Lijuan Chen, Huan Tao, Jian Huang, Chaoqun Xu, Lin Li, Shiwei Ma, Tian Tian, Wei Liu, Lichun Xue, and et al. 2016. "Impact of SNPs on Protein Phosphorylation Status in Rice (Oryza sativa L.)" International Journal of Molecular Sciences 17, no. 11: 1738. https://doi.org/10.3390/ijms17111738