Metabolic Response to XD14 Treatment in Human Breast Cancer Cell Line MCF-7
Abstract
:1. Introduction
2. Results
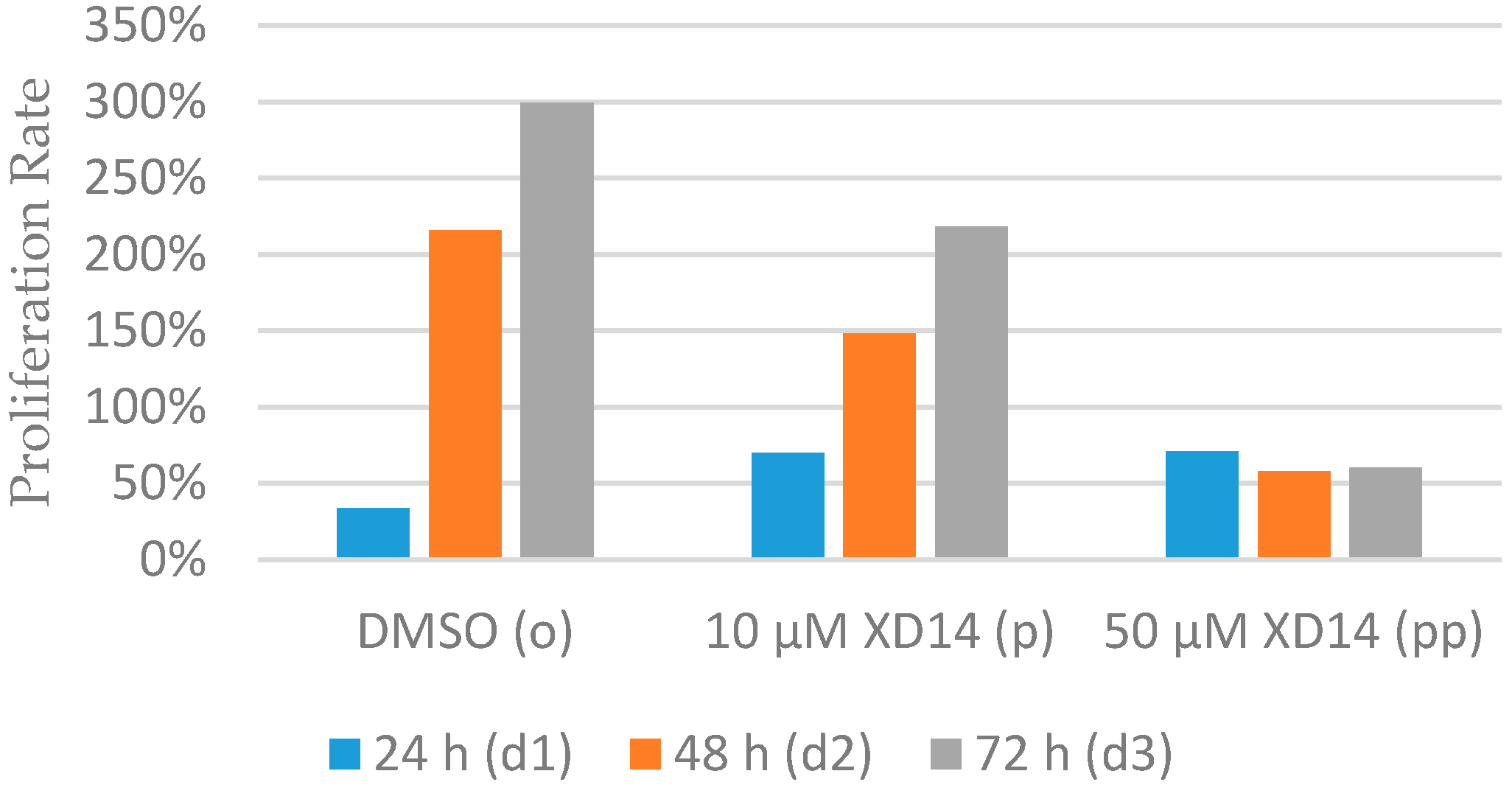
2.1. Inhibition of Cell Proliferation after Treatment with XD14
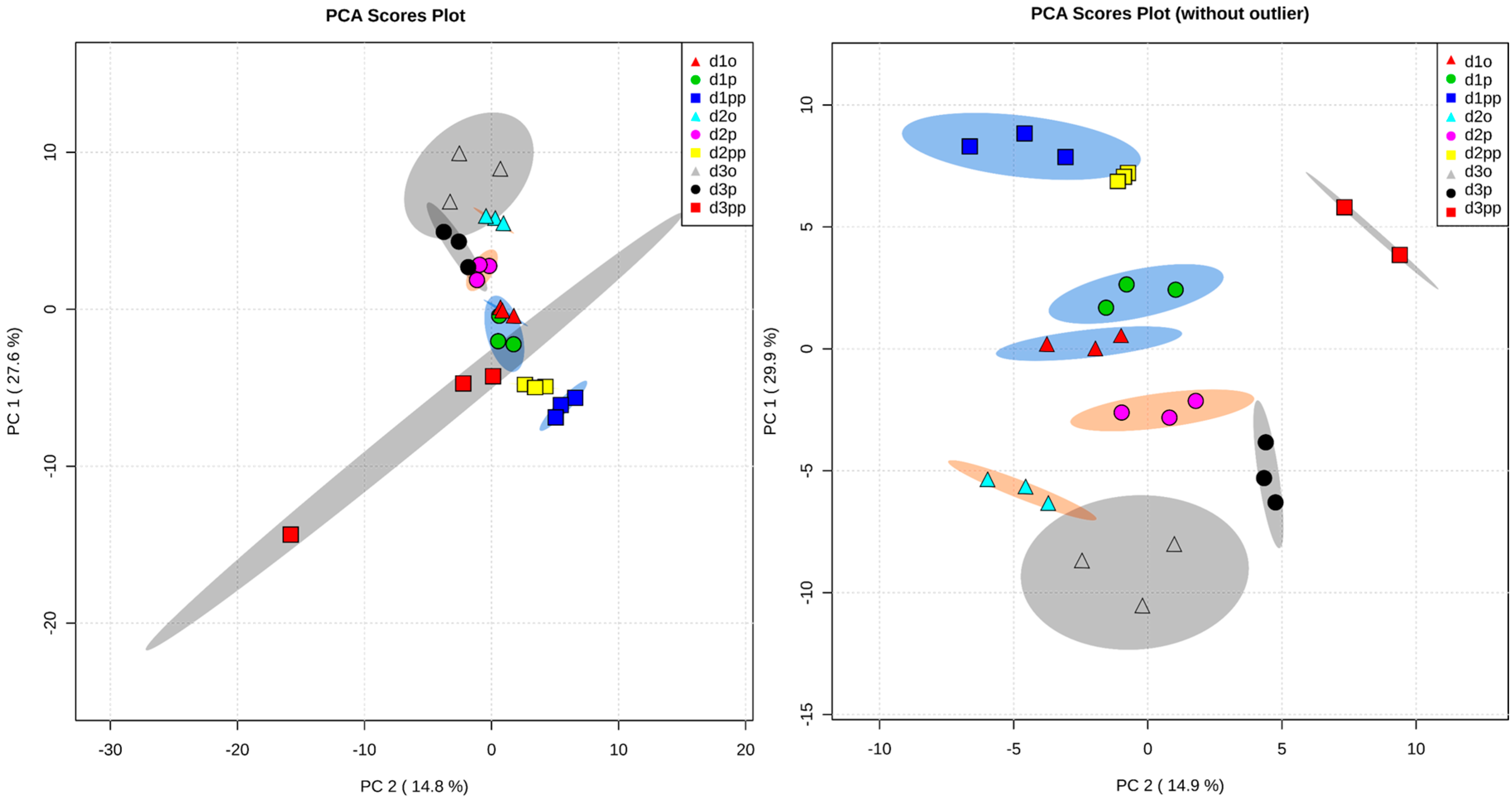
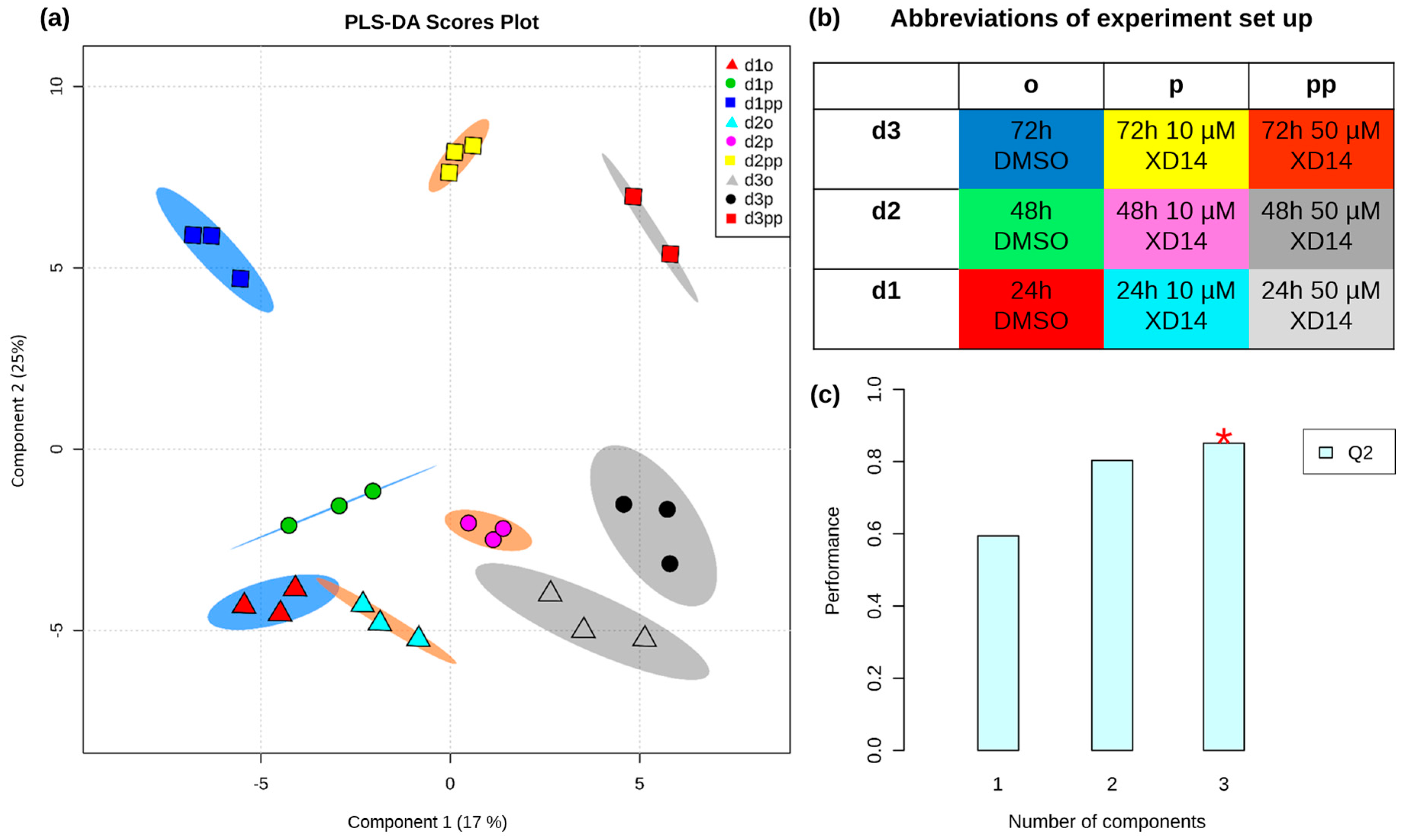
2.2. Global Metabolic Profile and Statistical Evaluation
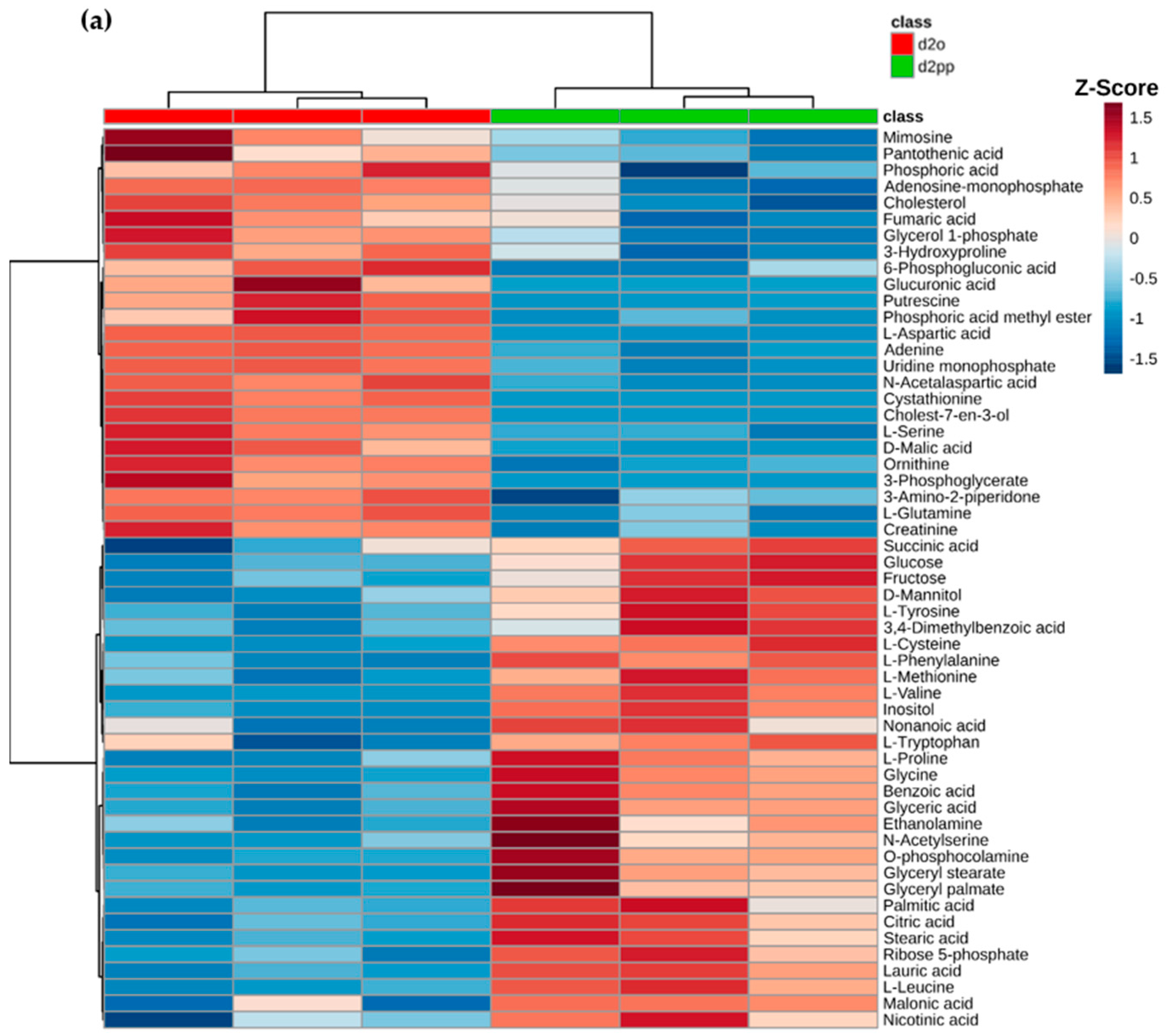
2.3. Alterations of Metabolic Profile after Two Days of 50 µM XD14 Treatment
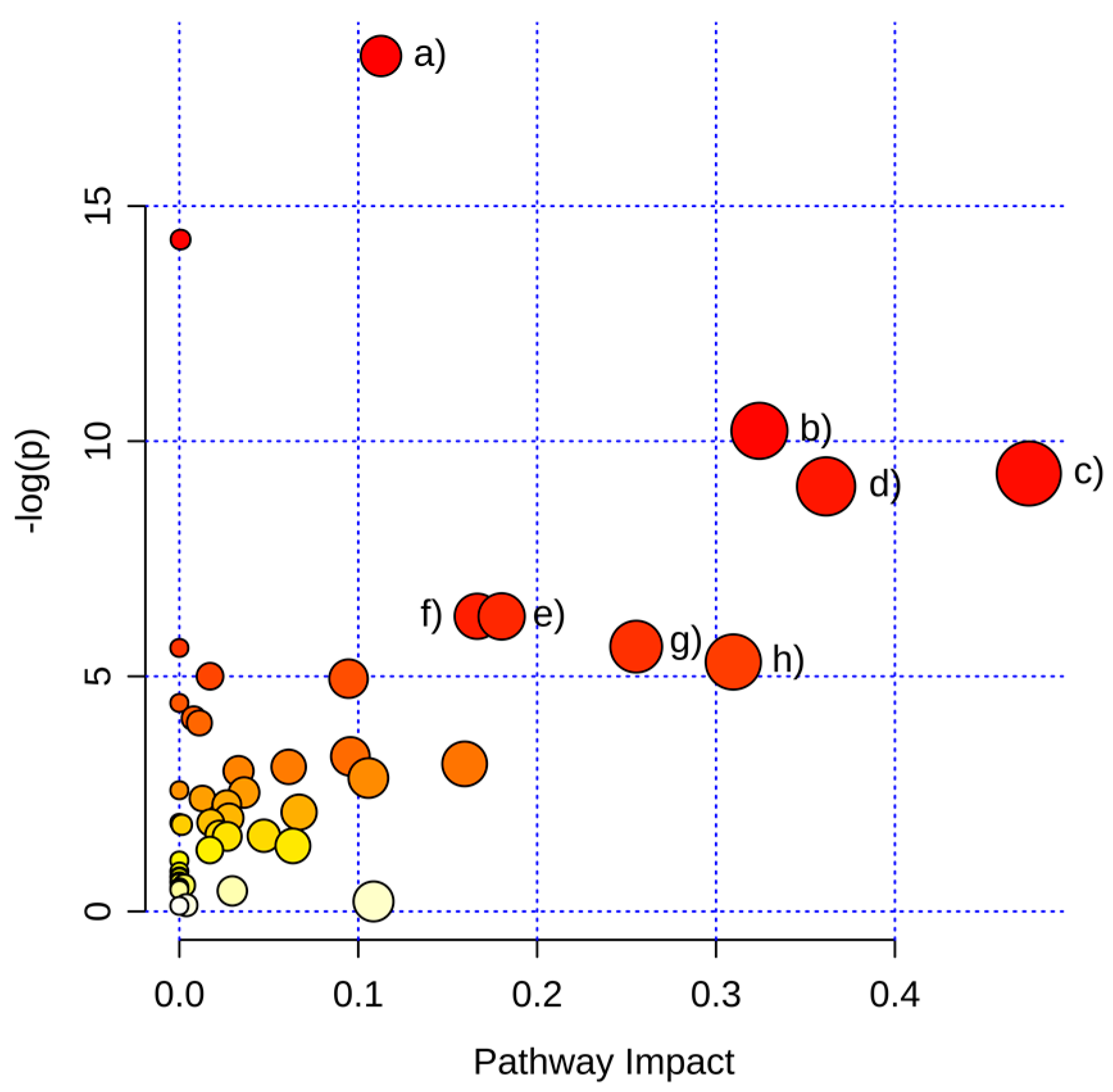
2.3.1. Pathway Analysis of Significantly Regulated Metabolites
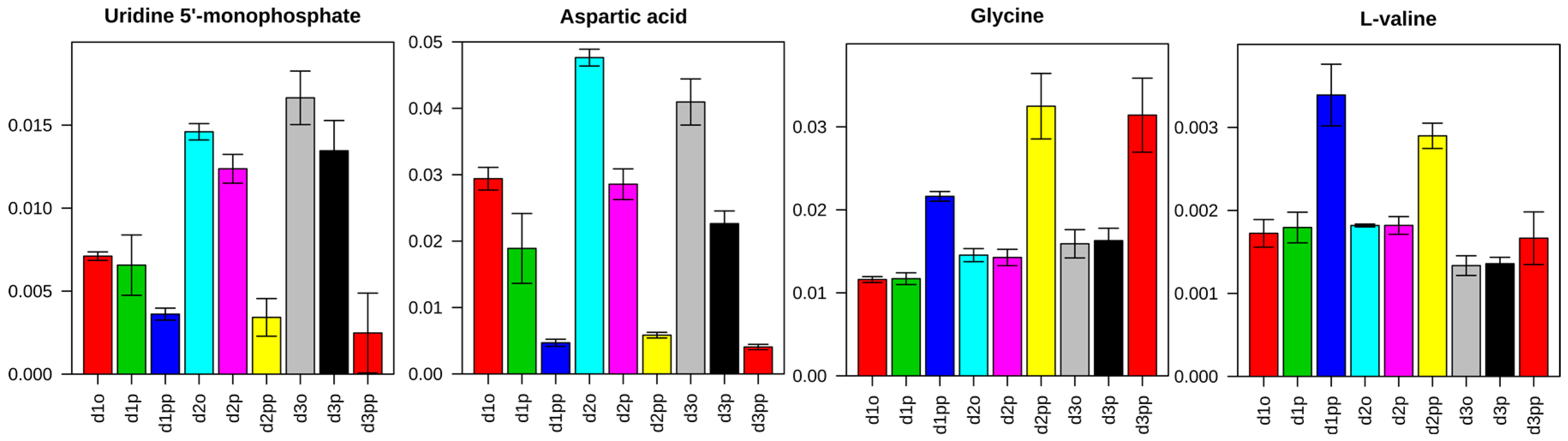
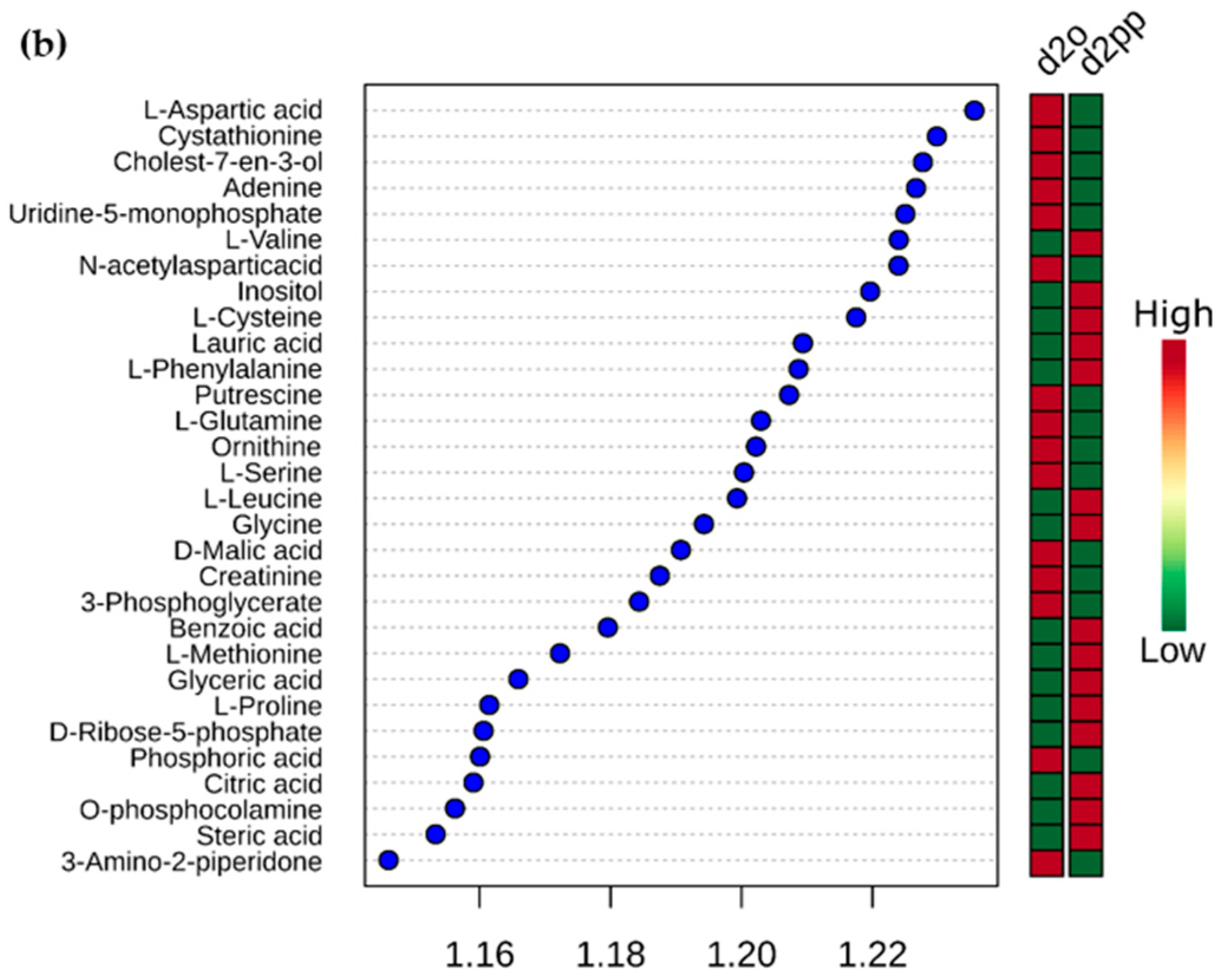
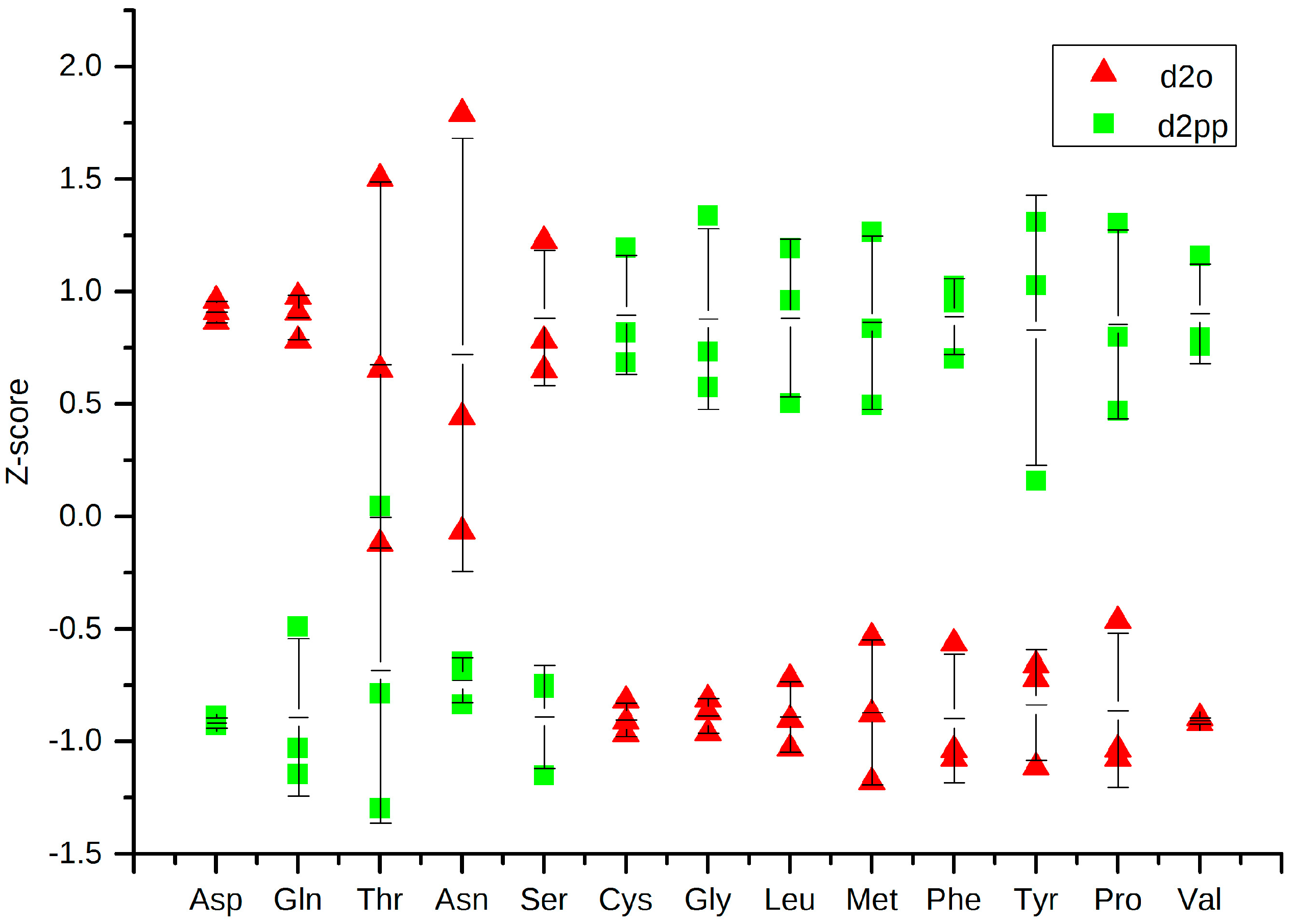
2.3.2. Change in Amino Acid Metabolism
2.3.3. Dysregulations of Other Metabolites
3. Discussion
4. Materials and Methods
4.1. Cell Culture and XD14 Treatment
4.2. Metabolite Extraction
4.3. Derivatization and GC-MS Analysis
4.4. Data Processing and Statistical Evaluation
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
| Group\Time | 0 h (d0) | 24 h (d1) | 48 h (d2) | 72 h (d3) |
|---|---|---|---|---|
| o | 1.33 × 106 | 1.78 × 106 | 4.20 × 106 | 5.31 × 106 |
| p | 1.33 × 106 | 2.26 × 106 | 3.30 × 106 | 4.23 × 106 |
| pp | 1.33 × 106 | 2.27 × 106 | 2.10 × 106 | 2.13 × 106 |
| Metabolites | p Value | FDR | Metabolites | p Value | FDR |
|---|---|---|---|---|---|
| Amino acid | Lipid/phospholipid and their derivatives | ||||
| l-Aspartic acid | 3.65 × 10−13 | 4.23 × 10−11 | Cholesterol | 4.39 × 10−4 | 1.18 × 10−3 |
| l-Serine | 5.94 × 10−8 | 6.89 × 10−7 | Cholest-7-en-3-ol | 2.62 × 10−9 | 3.79 × 10−8 |
| Glycine | 5.65 × 10−11 | 2.18 × 10−9 | Heptanoic acid | 5.83 × 10−5 | 2.05 × 10−4 |
| l-Leucine | 1.46 × 10−6 | 1.13 × 10−5 | Stearic acid | 2.85 × 10−4 | 8.26 × 10−4 |
| l-Methionine | 8.54 × 10−6 | 4.31 × 10−5 | Glyceryl stearate | 1.96 × 10−4 | 5.97 × 10−4 |
| l-Valine | 4.01 × 10−10 | 7.75 × 10−9 | Palmitic acid | 7.78 × 10−4 | 1.88 × 10−3 |
| l-Tyrosine | 4.41 × 10−6 | 2.56 × 10−5 | Glyceryl palmate | 1.53 × 10−3 | 3.42 × 10−3 |
| l-Tryptophan | 5.45 × 10−6 | 3.01 × 10−5 | Myristic acid | 5.48 × 10−3 | 1.01 × 10−2 |
| l-Glutamine | 5.46 × 10−5 | 1.98 × 10−4 | Lauric acid | 5.56 × 10−3 | 1.01 × 10−2 |
| l-Cysteine | 3.29 × 10−5 | 1.24 × 10−4 | Nonanoic acid | 9.14 × 10−3 | 1.45 × 10−2 |
| l-Phenylalanine | 2.30 × 10−4 | 6.83 × 10−4 | Palmitoleic acid | 8.26 × 10−3 | 1.39 × 10−2 |
| l-Threonine | 9.03 × 10−4 | 2.14 × 10−3 | Arachidic acid | 3.43 × 10−6 | 2.21 × 10−5 |
| l-Proline | 2.61 × 10−3 | 5.32 × 10−3 | O-phosphocolamine | 2.19 × 10−11 | 1.27 × 10−9 |
| l-Asparagine | 2.87 × 10−3 | 5.64 × 10−3 | Phosphoric acid | 7.76 × 10−4 | 1.88 × 10−3 |
| l-Glutamic acid | 9.92 × 10−5 | 3.20 × 10−4 | Phosphoric acid monomethyl ester | 5.62 × 10−4 | 1.48 × 10−3 |
| N-acetyl-Aspartic acid | 3.31 × 10−5 | 1.24 × 10−4 | Glycerol 1-phosphate | 8.38 × 10−3 | 1.39 × 10−2 |
| N-acetyl-Serine | 2.13 × 10−6 | 1.54 × 10−5 | Pyrophosphate | 5.90 × 10−3 | 1.05 × 10−2 |
| 4-hydroxy-Proline | 2.59 × 10−5 | 1.07 × 10−4 | Purine and Pyrimidine metabolism | ||
| Creatinine | 8.50 × 10−5 | 2.90 × 10−4 | Adenosine-5-monophosphate | 3.27 × 10−5 | 1.24 × 10−4 |
| Citrulline | 1.22 × 10−3 | 2.79 × 10−3 | Adenine | 2.17 × 10−3 | 4.67 × 10−3 |
| Ornithine | 2.53 × 10−6 | 1.73 × 10−5 | 5-deoxy-5-methylthio-Adenosine | 2.27 × 10−3 | 4.78 × 10−3 |
| 3-Amino-2-piperidone | 1.56 × 10−5 | 7.26 × 10−5 | Guanosine-5-monophosphate | 7.73 × 10−3 | 1.32 × 10−2 |
| Glycolysis and TCA intermediates | Uridine 5′-monophosphate | 2.03 × 10−10 | 5.00 × 10−9 | ||
| d-Glucose-6-phosphate | 1.73 × 10−5 | 7.45 × 10−5 | Others | ||
| d-Mannose-6-phosphate | 1.20 × 10−5 | 5.78 × 10−5 | Malonic acid | 1.73 × 10−5 | 7.45 × 10−5 |
| d-Gluconic acid | 9.51 × 10−10 | 1.58 × 10−8 | Aminomalonic acid | 6.90 × 10−4 | 1.74 × 10−3 |
| 3-Phosphoglycerate | 1.74 × 10−8 | 2.25 × 10−7 | Taurine | 5.06 × 10−3 | 9.63 × 10−3 |
| d-Malic acid | 8.62 × 10−8 | 8.39 × 10−7 | Pantothenic acid | 5.87 × 10−7 | 5.24 × 10−6 |
| Lactic acid | 1.35 × 10−6 | 1.12 × 10−5 | Threonic acid | 6.39 × 10−3 | 1.12 × 10−2 |
| Citric acid | 6.19 × 10−6 | 3.26 × 10−5 | Propanediol | 5.43 × 10−3 | 1.01 × 10−2 |
| Fumaric acid | 3.99 × 10−4 | 1.10 × 10−3 | d-Threitol | 8.84 × 10−5 | 2.93 × 10−4 |
| Glyceric acid | 6.68 × 10−4 | 1.72 × 10−3 | Inositol | 1.01 × 10−3 | 2.34 × 10−3 |
| Glycolic acid | 2.81 × 10−3 | 5.61 × 10−3 | Cystathionine | 2.15 × 10−10 | 5.00 × 10−9 |
| Glucose | 3.42 × 10−3 | 6.61 × 10−3 | Putrescine | 8.68 × 10−8 | 8.39 × 10−7 |
| Fructose | 8.51 × 10−3 | 1.39 × 10−2 | |||
| 6-Phosphogluconic acid | 4.31 × 10−6 | 2.56 × 10−5 | |||
| Order | Sample | Order | Sample | Order | Sample | Order | Sample |
|---|---|---|---|---|---|---|---|
| 1 | d3pp_1 | 8 | d2p_1 | 15 | d3o_2 | 22 | d3p_2 |
| 2 | d2pp_2 | 9 | d2o_2 | 16 | d1p_2 | 23 | d3pp_3 |
| 3 | d2pp_1 | 10 | d1pp_2 | 17 | d3p_1 | 24 | d2p_3 |
| 4 | d2pp_3 | 11 | d2o_3 | 18 | d3o_3 | 25 | d1o_3 |
| 5 | d1pp_1 | 12 | d3o_1 | 19 | d1p_3 | 26 | d1pp_3 |
| 6 | d1o_1 | 13 | d1p_1 | 20 | d3pp_2 | 27 | d3p_3 |
| 7 | d2o_1 | 14 | d2p_2 | 21 | d1o_2 |
| Pathways | Raw p | FDR | Impact |
|---|---|---|---|
| Aminoacyl-tRNA biosynthesis | 1.26 × 10−8 | 1.01 × 10−6 | 0.11 |
| Nitrogen metabolism | 6.24 × 10−7 | 2.50 × 10−5 | 0.00 |
| Glycine, serine and threonine metabolism | 3.64 × 10−5 | 9.72 × 10−4 | 0.32 |
| Alanine, aspartate and glutamate metabolism | 9.03 × 10−5 | 1.81 × 10−3 | 0.47 |
| Arginine and proline metabolism | 1.19 × 10−4 | 1.90 × 10−3 | 0.36 |
| Phenylalanine metabolism | 1.88 × 10−3 | 2.16 × 10−2 | 0.17 |
| Pantothenate and CoA biosynthesis | 1.89 × 10−3 | 2.16 × 10−2 | 0.18 |
| Pentose phosphate pathway | 3.59 × 10−3 | 3.28 × 10−2 | 0.26 |
| Cyanoamino acid metabolism | 3.69 × 10−3 | 3.28 × 10−2 | 0.00 |
| Cysteine and methionine metabolism | 4.97 × 10−3 | 3.98 × 10−2 | 0.31 |
| Glutathione metabolism | 6.73 × 10−3 | 4.72 × 10−2 | 0.02 |
| Citrate cycle (TCA cycle) | 7.09 × 10−3 | 4.72 × 10−2 | 0.09 |
References
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. 2013. Available online: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx (accessed on 2 March 2015).
- Howlader, N.; Noone, A.M.; Krapcho, M.; Garshell, J.; Miller, D.; Altekruse, S.F.; Kosary, C.L.; Yu, M.; Ruhl, J.; Tatalovich, Z.; et al. SEER Cancer Statistics Review, 1975–2011. Available online: http://seer.cancer.gov/csr/1975_2011 (accessed on 2 March 2015).
- Robert-Koch-Institut. Krebs in Deutschland 2009/2010. Available online: http://www.rki.de/DE/Content/Gesundheitsmonitoring/Gesundheitsberichterstattung/GBEDownloadsB/KID2013.pdf%3F__blob%3 DpublicationFile (accessed on 2 March 2015).
- Fan, L.; Strasser-Weippl, K.; Li, J.-J.; St Louis, J.; Finkelstein, D.M.; Yu, K.-D.; Chen, W.-Q.; Shao, Z.-M.; Goss, P.E. Breast cancer in China. Lancet Oncol. 2014, 15, e279–e289. [Google Scholar] [CrossRef]
- Lucas, X.; Wohlwend, D.; Hügle, M.; Schmidtkunz, K.; Gerhardt, S.; Schüle, R.; Jung, M.; Einsle, O.; Günther, S. 4-Acyl pyrroles: Mimicking acetylated lysines in histone code reading. Angew. Chem. Int. Ed. Engl. 2013, 52, 14055–14059. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.-P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Müller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Zhou, M.-M. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002, 513, 124–128. [Google Scholar] [CrossRef]
- Owen, D.J.; Ornaghi, P.; Yang, J.C.; Lowe, N.; Evans, P.R.; Ballario, P.; Neuhaus, D.; Filetici, P.; Travers, A.A. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase GCN5P. EMBO J. 2000, 19, 6141–6149. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Filippakopoulos, P.; Knapp, S. Bromodomains as therapeutic targets. Exp. Rev. Mol. Med. 2011, 13, e29. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Vakoc, C.R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 2014, 54, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wu, X.; Zhang, X.; Hua, W.; Zhang, Y.; Maimaiti, Y.; Gao, Z.; Zhang, Y. Inhibition of BRD4 suppresses tumor growth and enhances iodine uptake in thyroid cancer. Biochem. Biophys. Res. Commun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, F.; Jorge Moura Barbosa, A.; Daniele Parenti, M.; del Rio, A. Modulation of epigenetic targets for anticancer therapy: Clinicopathological relevance, structural data and drug discovery perspectives. Curr. Pharm. Des. 2013, 19, 578–613. [Google Scholar] [CrossRef] [PubMed]
- Di Costanzo, A.; del, G.N.; Migliaccio, A.; Altucci, L. Epigenetic drugs against cancer: An evolving landscape. Arch. Toxicol. 2014, 88, 1651–1668. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.I.; Robson, S.C.; Chung, C.W.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.W.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Raftery, D. Mass Spectrometry in Metabolomics; Springer: New York, NY, USA, 2014. [Google Scholar]
- Sana, T.R.; Gordon, D.B.; Fischer, S.M.; Tichy, S.E.; Kitagawa, N.; Lai, C.; Gosnell, W.L.; Chang, S.P. Global mass spectrometry based metabolomics profiling of erythrocytes infected with Plasmodium falciparum. PLoS ONE 2013, 8, e60840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.-H.; Sun, H.; Qiu, S.; Wang, X.-J. Metabolomics in noninvasive breast cancer. Clin. Chim. Acta 2013, 424, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar]
- Teresa, W.-M.F.; Richard, M.H.; Andrew, N.L. The Handbook of Metabolomics: Methods in Pharmacology and Toxicology; Humana Press; Springer Science: New York, NY, USA, 2012. [Google Scholar]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science (New York, N.Y.) 2012, 336, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Pfitzner, B.M.; Györffy, B.; Winzer, K.-J.; Radke, C.; Dietel, M.; Fiehn, O.; Denkert, C. Glutamate enrichment as new diagnostic opportunity in breast cancer. Int. J. Cancer 2015, 136, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Kabil, O.; Motl, N.; Banerjee, R. H2S and its role in redox signaling. Biochim. Biophys. Acta 2014, 1844, 1355–1366. [Google Scholar] [CrossRef] [PubMed]
- Liesenfeld, D.B.; Habermann, N.; Owen, R.W.; Scalbert, A.; Ulrich, C.M. Review of mass spectrometry-based metabolomics in cancer research. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2182–2201. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yan, L.; Liu, S.; Ambrosone, C.B.; Zhao, H. Plasma metabolomic profiles in breast cancer patients and healthy controls: By race and tumor receptor subtypes. Transl. Oncol. 2013, 6, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Willmann, L.; Schlimpert, M.; Halbach, S.; Erbes, T.; Stickeler, E.; Kammerer, B. Metabolic profiling of breast cancer: Differences in central metabolism between subtypes of breast cancer cell lines. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 1000, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Bullinger, D.; Neubauer, H.; Fehm, T.; Laufer, S.; Gleiter, C.H.; Kammerer, B. Metabolic signature of breast cancer cell line MCF-7: Profiling of modified nucleosides via LC-IT MS coupling. BMC Biochem. 2007, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Willmann, L.; Schlimpert, M.; Hirschfeld, M.; Erbes, T.; Neubauer, H.; Stickeler, E.; Kammerer, B. Alterations of the exo- and endometabolite profiles in breast cancer cell lines: A mass spectrometry-based metabolomics approach. Anal. Chim. Acta 2016, 925, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Frickenschmidt, A.; Frohlich, H.; Bullinger, D.; Zell, A.; Laufer, S.; Gleiter, C.H.; Liebich, H.; Kammerer, B. Metabonomics in cancer diagnosis: Mass spectrometry-based profiling of urinary nucleosides from breast cancer patients. Biomarkers 2008, 13, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.E. An integrated method for spectrum extraction and compound identification from GC/MS data. J. Am. Soc. Mass Spectrom. 1999, 10, 770–781. [Google Scholar] [CrossRef]
- Styczynski, M.P.; Moxley, J.F.; Tong, L.V.; Walther, J.L.; Jensen, K.L.; Stephanopoulos, G.N. Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Anal. Chem. 2007, 79, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef] [PubMed]
- Hugle, M.; Lucas, X.; Weitzel, G.; Ostrovskyi, D.; Breit, B.; Gerhardt, S.; Einsle, O.; Gunther, S.; Wohlwend, D. 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1). J. Med. Chem. 2016, 59, 1518–1530. [Google Scholar] [CrossRef] [PubMed]
- Labuschagne, C.F.; van den Broek, N.J.F.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D.K. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Gruber, C.J.; Tschugguel, W.; Schneeberger, C.; Huber, J.C. Production and actions of estrogens. N. Engl. J. Med. 2002, 346, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Key, T.; Appleby, P.; Barnes, I.; Reeves, G. Endogenous sex hormones and breast cancer in postmenopausal women reanalysis of nine prospective studies. J. Natl. Cancer Inst. 2002, 94. [Google Scholar] [CrossRef]
- Fournier, A.; Berrino, F.; Riboli, E.; Avenel, V.; Clavel-Chapelon, F. Breast cancer risk in relation to different types of hormone replacement therapy in the E3N-EPIC cohort. Int. J. Cancer 2005, 114, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Milovic, V.; Stein, J.; Odera, G.; Gilani, S.; Murphy, G.M. Low-dose deoxycholic acid stimulates putrescine uptake in colon cancer cells (Caco-2). Cancer Lett. 2000, 154, 195–200. [Google Scholar] [CrossRef]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef] [PubMed]
- Takao, K.; Rickhag, M.; Hegardt, C.; Oredsson, S.; Persson, L. Induction of apoptotic cell death by putrescine. Int. J. Biochem. Cell Biol. 2006, 38, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Schneider, Y.; Chabert, P.; Stutzmann, J.; Coelho, D.; Fougerousse, A.; Gossé, F.; Launay, J.-F.; Brouillard, R.; Raul, F. Resveratrol analog (Z)-3,5,4′-trimethoxystilbene is a potent anti-mitotic drug inhibiting tubulin polymerization. Int. J. Cancer 2003, 107, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolite Profiling in Arabidopsis. Methods Mol. Biol. 2006, 323, 439–447. [Google Scholar]
- NIST/EPA/NI. NIST Mass Spectral Libra 2011. Available online: http://www.sisweb.com/software/ms/nist.htm (accessed on 15 July 2015).
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmuller, E.; Dormann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. [email protected]: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Ana. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, D.; Kather, M.; Willmann, L.; Schlimpert, M.; Bauer, C.; Lagies, S.; Schmidtkunz, K.; Eisenhardt, S.U.; Jung, M.; Günther, S.; et al. Metabolic Response to XD14 Treatment in Human Breast Cancer Cell Line MCF-7. Int. J. Mol. Sci. 2016, 17, 1772. https://doi.org/10.3390/ijms17101772
Pan D, Kather M, Willmann L, Schlimpert M, Bauer C, Lagies S, Schmidtkunz K, Eisenhardt SU, Jung M, Günther S, et al. Metabolic Response to XD14 Treatment in Human Breast Cancer Cell Line MCF-7. International Journal of Molecular Sciences. 2016; 17(10):1772. https://doi.org/10.3390/ijms17101772
Chicago/Turabian StylePan, Daqiang, Michel Kather, Lucas Willmann, Manuel Schlimpert, Christoph Bauer, Simon Lagies, Karin Schmidtkunz, Steffen U. Eisenhardt, Manfred Jung, Stefan Günther, and et al. 2016. "Metabolic Response to XD14 Treatment in Human Breast Cancer Cell Line MCF-7" International Journal of Molecular Sciences 17, no. 10: 1772. https://doi.org/10.3390/ijms17101772