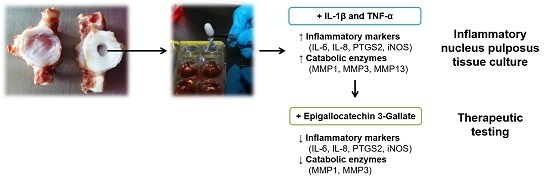
An Inflammatory Nucleus Pulposus Tissue Culture Model to Test Molecular Regenerative Therapies: Validation with Epigallocatechin 3-Gallate
Abstract
:1. Introduction
2. Results
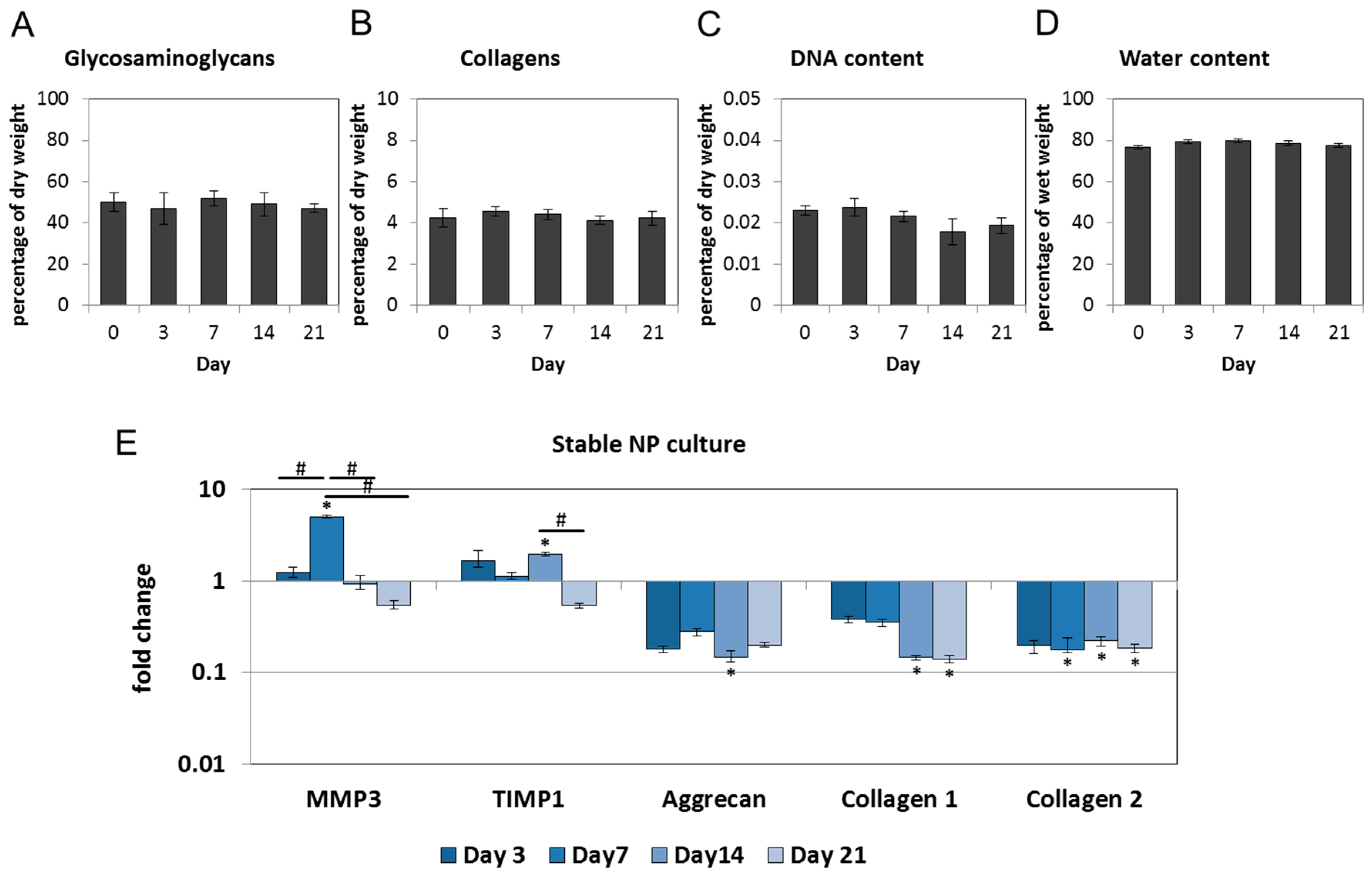
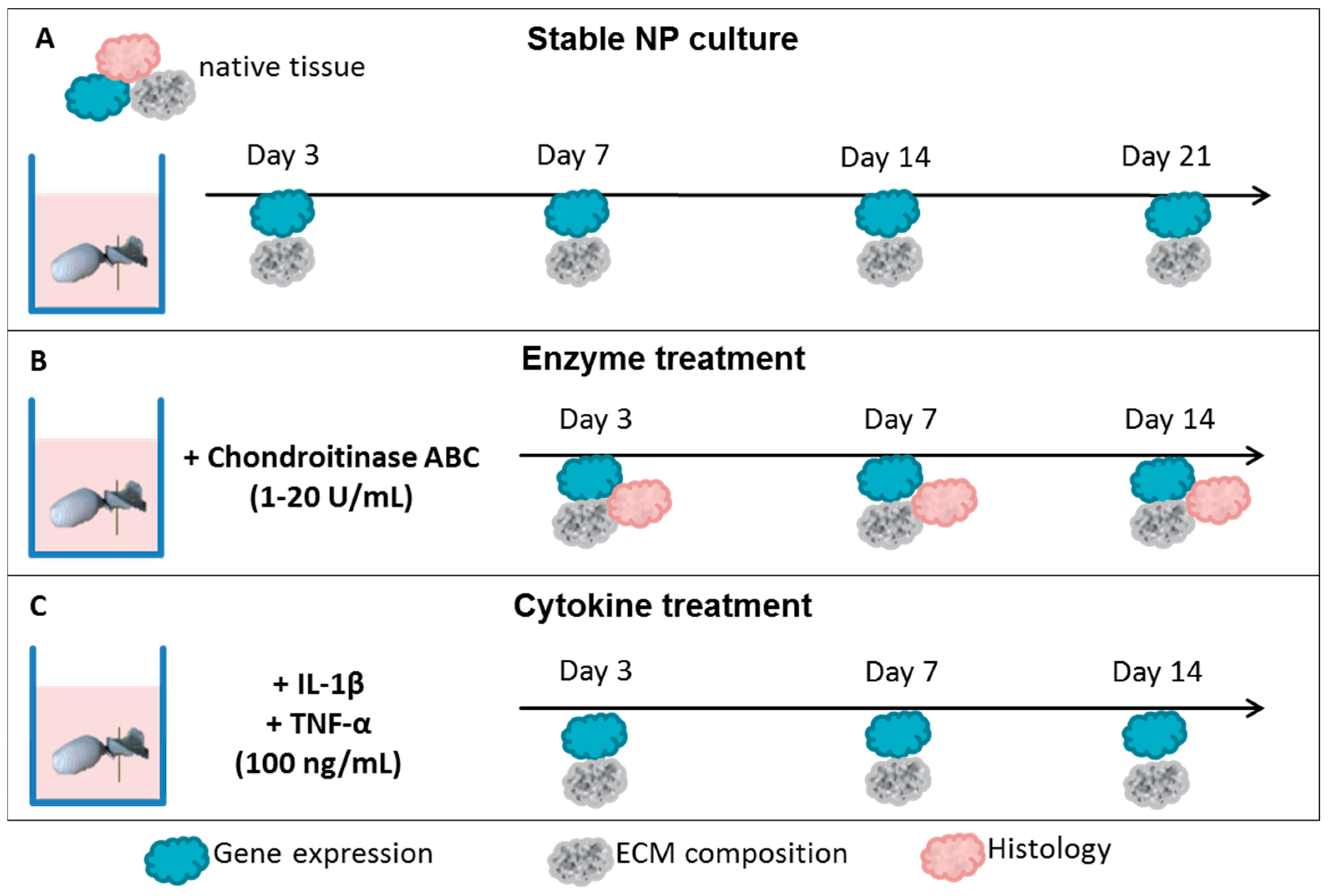
2.1. Nucleus Pulposus Tissue in Dyneema Jackets Is Stable over a Period of 21 Days
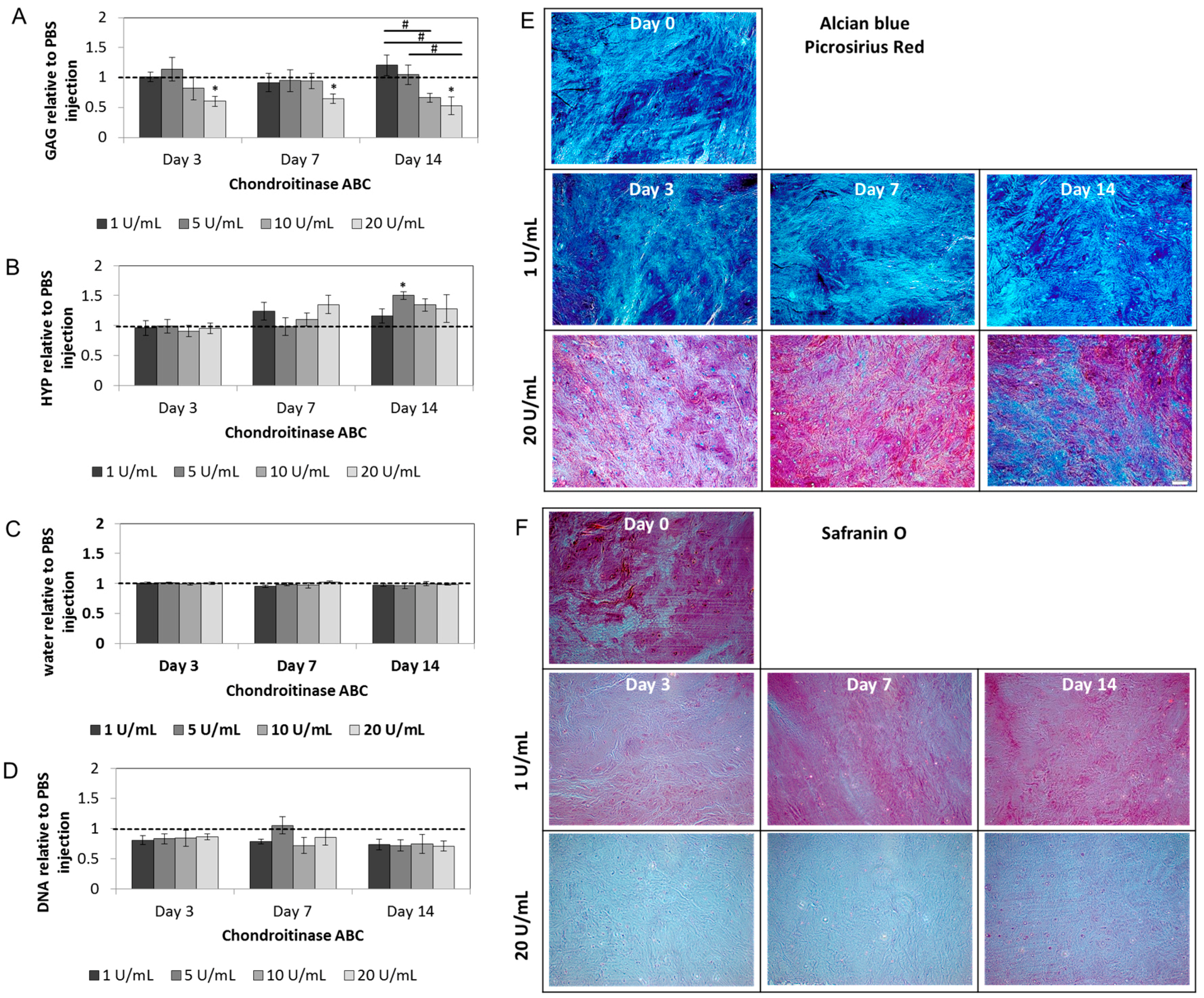
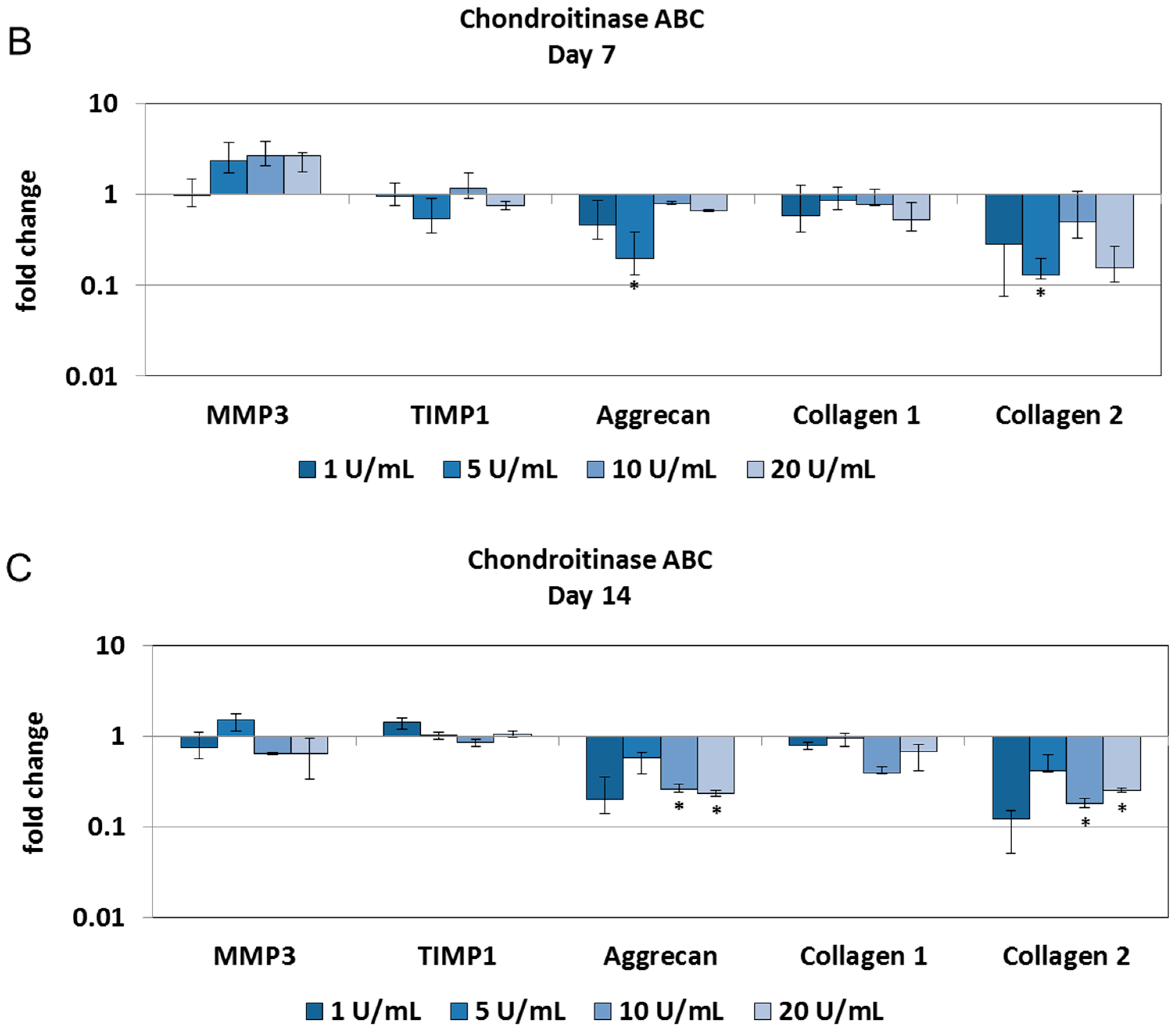
2.2. Chondroitinase ABC Reduces Proteoglycan Content in the NP but Does Not Induce Cell Responses
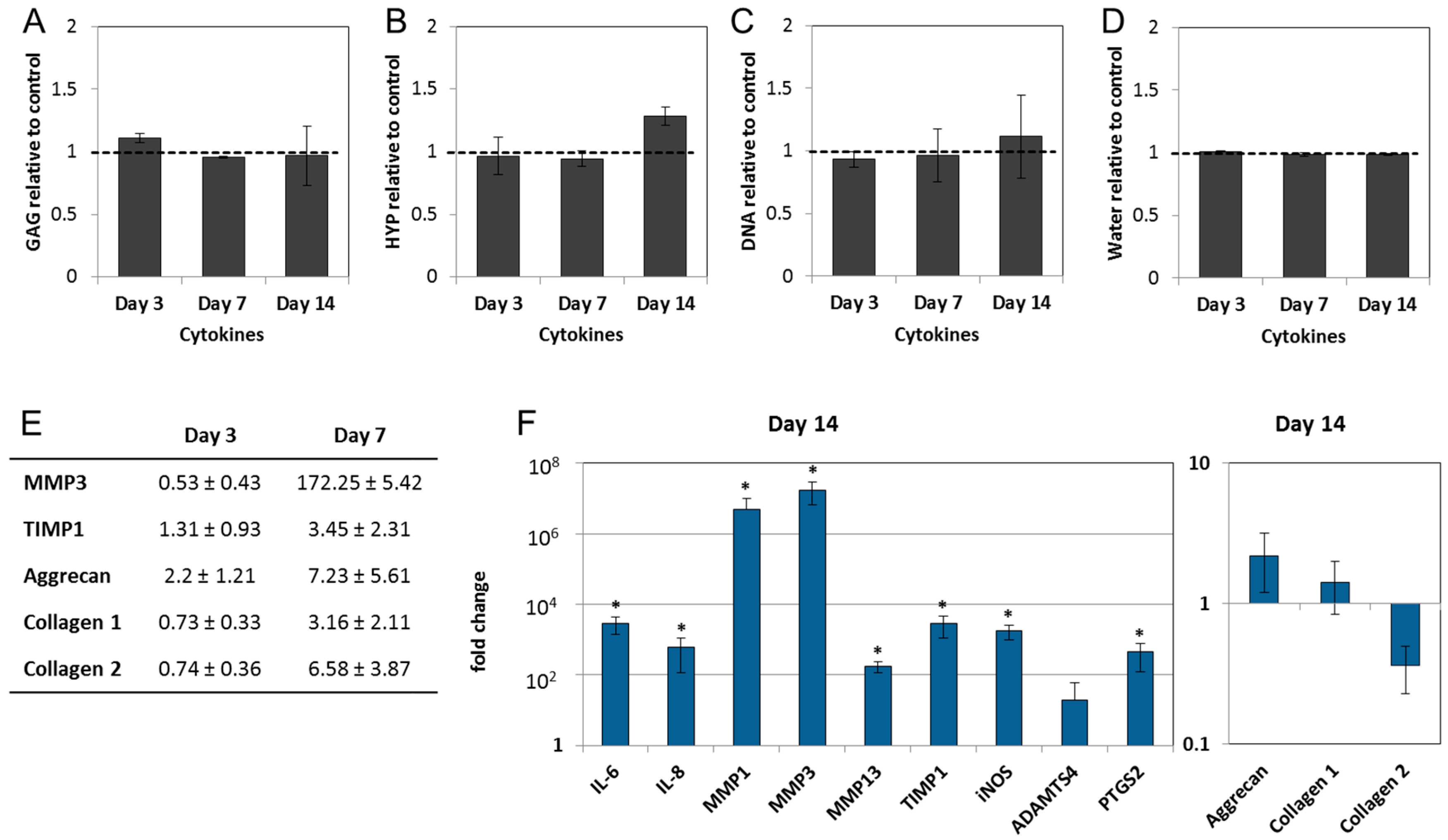
2.3. Interleukin-1 Beta (IL-1β) and Tumor Necrosis Factor Alpha (TNF-α) Induce Gene Expression of Inflammatory and Pain-Related Markers in the NP Cultures
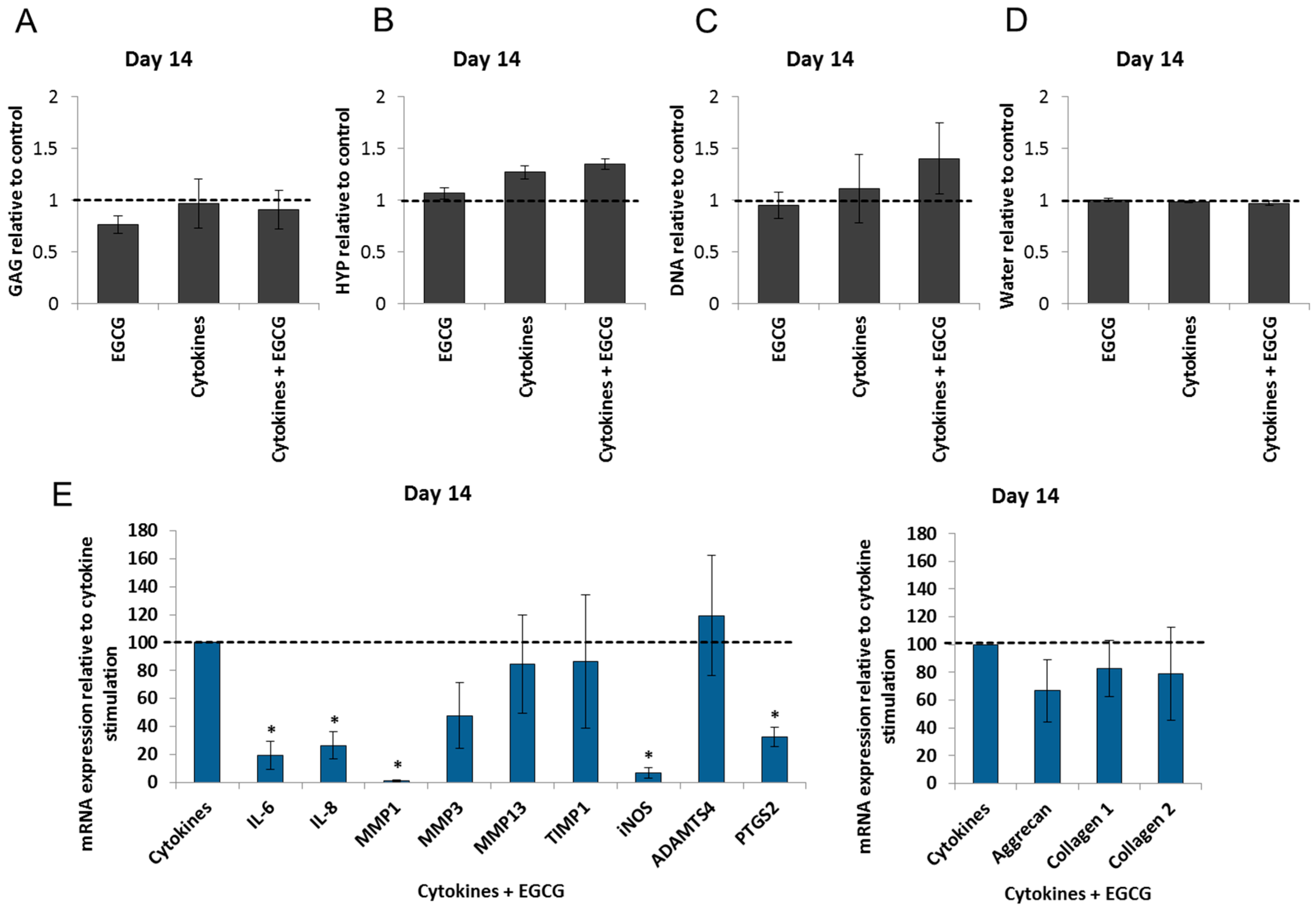
2.4. Epigallocatechin 3-Gallate Ameliorates the Inflammatory and Catabolic Phenotype in the NP Cultures
3. Discussion
4. Materials and Methods
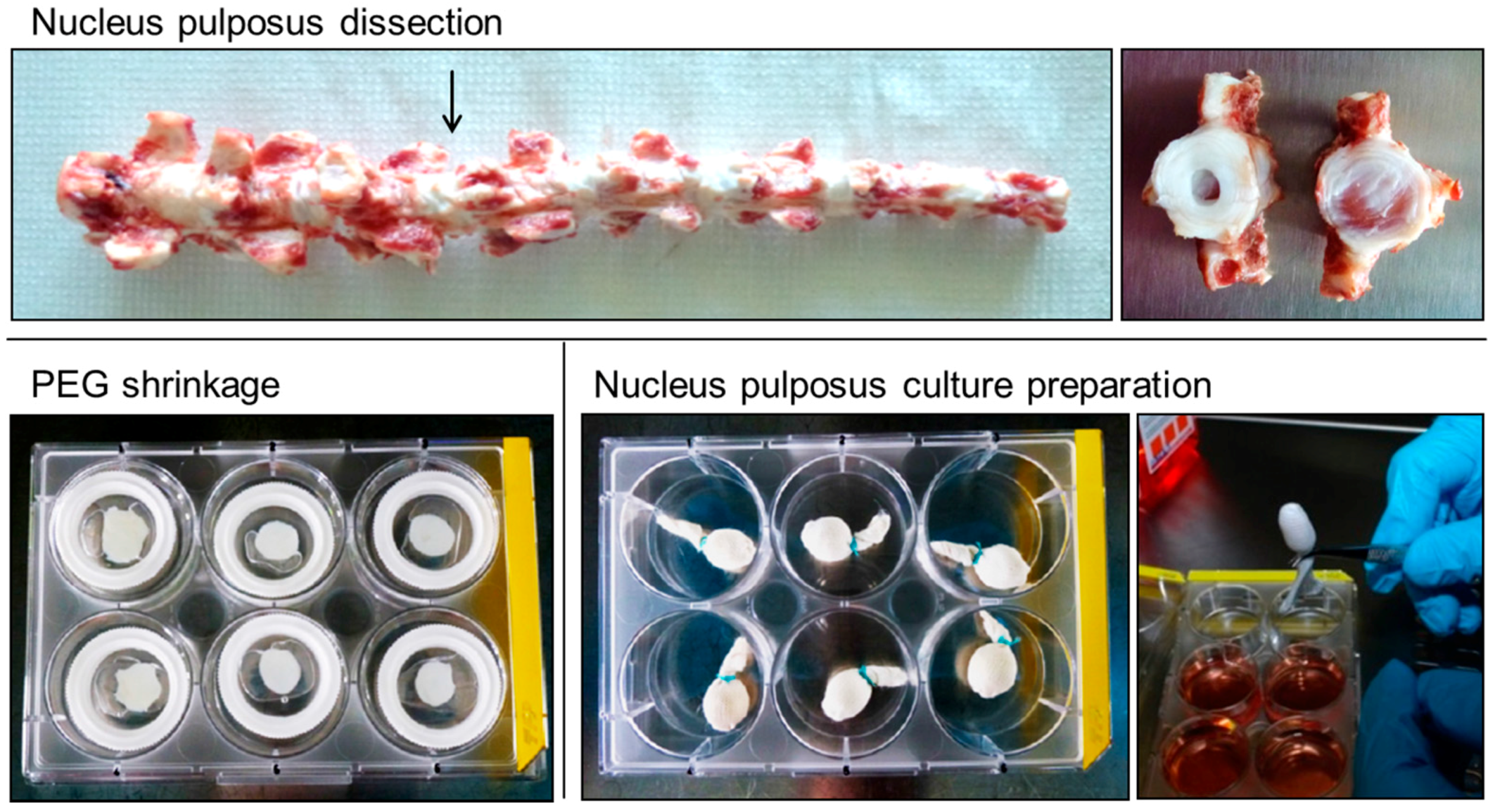
4.1. Bovine Nucleus Pulposus Tissue Culture
4.2. Induction of Nucleus Pulposus Degeneration
4.3. RNA Isolation
4.4. Gene Expression
4.5. Extracellular Matrix Composition (Glycosaminoglycans, Hydroxyproline, DNA, and Water)
4.6. Histology
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Urban, J.P.G.; Smith, S.; Fairbank, J.C.T. Nutrition of the intervertebral disc. Spine 2004, 29, 2700–2709. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.M.; Mobasheri, A.; Freemont, A.J.; Hoyland, J.A. Intervertebral disc biology, degeneration and novel tissue engineering and regenerative medicine therapies. Histol. Histopathol. 2007, 22, 1033–1041. [Google Scholar] [PubMed]
- Johnson, Z.I.; Schoepflin, Z.R.; Choi, H.; Shapiro, I.M.; Risbud, M.V. Disc in flames: Roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur. Cells Mater. 2015, 30, 104–117. [Google Scholar]
- Le Maitre, C.L.; Freemont, A.J.; Hoyland, J.A. Accelerated cellular senescence in degenerate intervertebral discs: A possible role in the pathogenesis of intervertebral disc degeneration. Arthritis Res. Ther. 2007, 9, R45. [Google Scholar] [CrossRef] [PubMed]
- Le Maitre, C.L.; Freemont, A.J.; Hoyland, J.A. The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res. Ther. 2005, 7, R732–R745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Maitre, C.L.; Hoyland, J.A.; Freemont, A.J. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1β and TNF-α expression profile. Arthritis Res. Ther. 2007, 9, R77. [Google Scholar] [CrossRef] [PubMed]
- Purmessur, D.; Walter, B.A.; Roughley, P.J.; Laudier, D.M.; Hecht, A.C.; Iatridis, J. A role for TNF-α in intervertebral disc degeneration: A non-recoverable catabolic shift. Biochem. Biophys. Res. Commun. 2013, 433, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Oda, H.; Matsuzaki, H.; Tokuhashi, Y.; Wakabayashi, K.; Uematsu, Y.; Iwahashi, M. Degeneration of intervertebral discs due to smoking: Experimental assessment in a rat-smoking model. J. Orthop. Sci. 2004, 9, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, B.; Potier, E.; van Dijk, M.; Langelaan, M.; Papen-Botterhuis, N.; Ito, K. Reduced tonicity stimulates an inflammatory response in nucleus pulposus tissue that can be limited by a COX-2-specific inhibitor. J. Orthop. Res. 2015, 33, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Vo, N.V.; Hartman, R.A.; Yurube, T.; Jacobs, L.J.; Sowa, G.A.; Kang, J.D. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J. 2013, 13, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Krupkova, O.; Sekiguchi, M.; Klasen, J.; Hausmann, O.; Konno, S.; Ferguson, S.J.; Wuertz-Kozak, K. Epigallocatechin 3-gallate suppresses interleukin-1β-induced inflammatory responses in intervertebral disc cells in vitro and reduces radiculopathic pain in rats. Eur. Cells Mater. 2014, 28, 372–386. [Google Scholar]
- Wuertz, K.; Haglund, L. Inflammatory mediators in intervertebral disk degeneration and discogenic pain. Glob. Spine J. 2013, 3, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Risbud, M.V.; Shapiro, I.M. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat. Rev. Rheumatol. 2014, 10, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Freemont, A.J.; Hoyland, J.A.; Rajpura, A.; Byers, R.J.; Bartley, C.; Jeziorska, M.; Knight, M.; Ross, R.; O’Brien, J.; Sutcliffe, J.; et al. Matrix degradation by chondrocytes in the intervertebral disc is mediated by interleukin-1. Rheumatology 2001, 40, 75. [Google Scholar]
- Wang, W.J.; Yu, X.H.; Wang, C.; Yang, W.; He, W.S.; Zhang, S.J.; Yan, Y.G.; Zhang, J. MMPs and ADAMTSs in intervertebral disc degeneration. Clin. Chim. Acta Int. J. Clin. Chem. 2015, 448, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.A.; Roughley, P.J. What is intervertebral disc degeneration, and what causes it? Spine 2006, 31, 2151–2161. [Google Scholar] [CrossRef] [PubMed]
- Galbusera, F.; van Rijsbergen, M.; Ito, K.; Huyghe, J.M.; Brayda-Bruno, M.; Wilke, H.J. Ageing and degenerative changes of the intervertebral disc and their impact on spinal flexibility. Eur. Spine J. 2014, 23 (Suppl. S3), S324–S332. [Google Scholar] [CrossRef] [PubMed]
- Kuslich, S.D.; Ulstrom, C.L.; Michael, C.J. The tissue origin of low back pain and sciatica: A report of pain response to tissue stimulation during operations on the lumbar spine using local anesthesia. Orthop. Clin. N. Am. 1991, 22, 181–187. [Google Scholar]
- Freemont, A.J.; Peacock, T.E.; Goupille, P.; Hoyland, J.A.; O’Brien, J.; Jayson, M.I.V. Nerve ingrowth into diseased intervertebral disc in chronic back pain. Lancet 1997, 350, 178–181. [Google Scholar] [CrossRef]
- Wang, Y.; Videman, T.; Battie, M.C. Issls prize winner: Lumbar vertebral endplate lesions associations with disc degeneration and back pain history. Spine 2012, 37, 1490–1496. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.; Sundaram, L.J. Low back pain: An economic assessment in the United States. Orthop. Clin. N. Am. 2004, 35, 1–5. [Google Scholar] [CrossRef]
- Jacobs, W.C.; Rubinstein, S.M.; Koes, B.; van Tulder, M.W.; Peul, W.C. Evidence for surgery in degenerative lumbar spine disorders. Best Pract. Res. Clin. Rheumatol. 2013, 27, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Verma, K.; Gandhi, S.D.; Maltenfort, M.; Albert, T.J.; Hilibrand, A.S.; Vaccaro, A.R.; Radcliff, K.E. Rate of adjacent segment disease in cervical disc arthroplasty versus single-level fusion: Meta-analysis of prospective studies. Spine 2013, 38, 2253–2257. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.C.; Gantenbein-Ritter, B. Intervertebral disc regeneration or repair with biomaterials and stem cell therapy—Feasible or fiction? Swiss Med. Wkly. 2012, 142, w13598. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D.; Grad, S. Advancing the cellular and molecular therapy for intervertebral disc disease. Adv. Drug Deliv. Rev. 2015, 84, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D.; Andersson, G.B. Stem cell therapy for intervertebral disc regeneration: Obstacles and solutions. Nat. Rev. Rheumatol. 2015, 11, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Illien-Junger, S.; Grosjean, F.; Laudier, D.M.; Vlassara, H.; Striker, G.E.; Iatridis, J.C. Combined anti-inflammatory and anti-age drug treatments have a protective effect on intervertebral discs in mice with diabetes. PLoS ONE 2013, 8, e64302. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Z.; Rui, Y.F.; Tan, Q.; Wang, C. Enhancing intervertebral disc repair and regeneration through biology: Platelet-rich plasma as an alternative strategy. Arthritis Res. Ther. 2013, 15, 220. [Google Scholar] [CrossRef] [PubMed]
- Sawamura, K.; Ikeda, T.; Nagae, M.; Okamoto, S.; Mikami, Y.; Hase, H.; Ikoma, K.; Yamada, T.; Sakamoto, H.; Matsuda, K.; et al. Characterization of in vivo effects of platelet-rich plasma and biodegradable gelatin hydrogel microspheres on degenerated intervertebral discs. Tissue Eng. Part A 2009, 15, 3719–3727. [Google Scholar] [CrossRef] [PubMed]
- Gawri, R.; Antoniou, J.; Ouellet, J.; Awwad, W.; Steffen, T.; Roughley, P.; Haglund, L.; Mwale, F. Best paper NASS 2013: Link-N can stimulate proteoglycan synthesis in the degenerated human intervertebral discs. Eur. Cells Mater. 2013, 26, 107–119. [Google Scholar]
- Mwale, F. Molecular therapy for disk degeneration and pain. Glob. Spine J. 2013, 3, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.J.; Ito, K.; Nolte, L.P. Fluid flow and convective transport of solutes within the intervertebral disc. J. Biomech. 2004, 37, 213–221. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Kannaiyan, R.; Sethi, G. Targeting cell signaling and apoptotic pathways by dietary agents: Role in the prevention and treatment of cancer. Nutr. Cancer 2011, 63, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Wuertz, K.; Quero, L.; Sekiguchi, M.; Klawitter, M.; Nerlich, A.; Konno, S.; Kikuchi, S.; Boos, N. The red wine polyphenol resveratrol shows promising potential for the treatment of nucleus pulposus-mediated pain in vitro and in vivo. Spine 2011, 36, E1373–E1384. [Google Scholar] [CrossRef] [PubMed]
- Klawitter, M.; Quero, L.; Klasen, J.; Gloess, A.N.; Klopprogge, B.; Hausmann, O.; Boos, N.; Wuertz, K. Curcuma DMSO extracts and curcumin exhibit an anti-inflammatory and anti-catabolic effect on human intervertebral disc cells, possibly by influencing TLR2 expression and JNK activity. J. Inflamm. 2012, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Klawitter, M.; Quero, L.; Klasen, J.; Liebscher, T.; Nerlich, A.; Boos, N.; Wuertz, K. Triptolide exhibits anti-inflammatory, anti-catabolic as well as anabolic effects and suppresses TLR expression and MAPK activity in IL-1β treated human intervertebral disc cells. Eur. Spine J. 2012, 21 (Suppl. S6), S850–S859. [Google Scholar] [CrossRef] [PubMed]
- Krupkova, O.; Handa, J.; Hlavna, M.; Klasen, J.; Ospelt, C.; Ferguson, S.J.; Wuertz-Kozak, K. The natural polyphenol epigallocatechin gallate protects intervertebral disc cells from oxidative stress. Oxid. Med. Cell. Longev. 2016, 2016, 7031397. [Google Scholar] [CrossRef] [PubMed]
- Vita, J.A. Polyphenols and cardiovascular disease: Effects on endothelial and platelet function. Am. J. Clin. Nutr. 2005, 81, 292S–297S. [Google Scholar] [PubMed]
- Gantenbein, B.; Illien-Junger, S.; Chan, S.C.W.; Walser, J.; Haglund, L.; Ferguson, S.J.; Iatridis, J.C.; Grad, S. Organ culture bioreactors—Platforms to study human intervertebral disc degeneration and regenerative therapy. Curr. Stem Cell Res. Ther. 2015, 10, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Gawri, R.; Mwale, F.; Ouellet, J.; Roughley, P.J.; Steffen, T.; Antoniou, J.; Haglund, L. Development of an organ culture system for long-term survival of the intact human intervertebral disc. Spine 2011, 36, 1835–1842. [Google Scholar] [CrossRef] [PubMed]
- Jim, B.; Steffen, T.; Moir, J.; Roughley, P.; Haglund, L. Development of an intact intervertebral disc organ culture system in which degeneration can be induced as a prelude to studying repair potential. Eur. Spine J. 2011, 20, 1244–1254. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.C.W.; Burki, A.; Bonel, H.M.; Benneker, L.M.; Gantenbein-Ritter, B. Papain-induced in vitro disc degeneration model for the study of injectable nucleus pulposus therapy. Spine J. 2013, 13, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Bucher, C.; Gazdhar, A.; Benneker, L.M.; Geiser, T.; Gantenbein-Ritter, B. Nonviral gene delivery of growth and differentiation factor 5 to human mesenchymal stem cells injected into a 3D bovine intervertebral disc organ culture system. Stem Cells Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Dudli, S.; Haschtmann, D.; Ferguson, S.J. Fracture of the vertebral endplates, but not equienergetic impact load, promotes disc degeneration in vitro. J. Orthop. Res. 2012, 30, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, M.A.A.; Ulrich, J.A.; Bass, E.C.; Rodriguez, A.G.; Liu, J.J.; Lotz, J.C. Stab incision for inducing intervertebral disc degeneration in the rat. Spine 2007, 32, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.C.; Liu, J.W.; Tang, S.Y. Longitudinal changes in the structure and inflammatory response of the intervertebral disc due to stab injury in a murine organ culture model. J. Orthop. Res. 2016, 34, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Markova, D.Z.; Kepler, C.K.; Addya, S.; Murray, H.B.; Vaccaro, A.R.; Shapiro, I.M.; Anderson, D.G.; Albert, T.J.; Risbud, M.V. An organ culture system to model early degenerative changes of the intervertebral disc II: Profiling global gene expression changes. Arthritis Res. Ther. 2013, 15, R121. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, G.Q.; Boldt, A.; Nagl, I.; Pereira, C.L.; Benz, K.; Wilke, H.J.; Ignatius, A.; Barbosa, M.A.; Goncalves, R.M.; Neidlinger-Wilke, C. A degenerative/proinflammatory intervertebral disc organ culture: An ex vivo model for anti-inflammatory drug and cell therapy. Tissue Eng. 2016, 22, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, B.G.; Potier, E.; Ito, K. Long-term culture of bovine nucleus pulposus explants in a native environment. Spine J. 2013, 13, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Wuertz, K.; Vo, N.; Kletsas, D.; Boos, N. Inflammatory and catabolic signalling in intervertebral discs: The roles of NF-κB and map kinases. Eur. Cells Mater. 2012, 23, 103–119. [Google Scholar]
- Alini, M.; Eisenstein, S.M.; Ito, K.; Little, C.; Kettler, A.A.; Masuda, K.; Melrose, J.; Ralphs, J.; Stokes, I.; Wilke, H.J. Are animal models useful for studying human disc disorders/degeneration? Eur. Spine J. 2008, 17, 2–19. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.A.; van Doeselaar, M.; Meij, B.P.; Tryfonidou, M.A.; Ito, K. The stimulatory effect of notochordal cell-conditioned medium in a nucleus pulposus explant culture. Tissue Eng. 2016, 22, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, B.G.; Potier, E.; van Dijk, M.; Creemers, L.B.; Ito, K. Osteogenic protein 1 does not stimulate a regenerative effect in cultured human degenerated nucleus pulposus tissue. J. Tissue Eng. Regen. Med. 2015. [Google Scholar] [CrossRef] [PubMed]
- Furtwangler, T.; Chan, S.C.W.; Bahrenberg, G.; Richards, P.J.; Gantenbein-Ritter, B. Assessment of the matrix degenerative effects of MMP-3, ADAMTS-4, and HTRA1, injected into a bovine intervertebral disc organ culture model. Spine 2013, 38, E1377–E1387. [Google Scholar] [CrossRef] [PubMed]
- Tiaden, A.N.; Klawitter, M.; Lux, V.; Mirsaidi, A.; Bahrenberg, G.; Glanz, S.; Quero, L.; Liebscher, T.; Wuertz, K.; Ehrmann, M.; et al. Detrimental role for human high temperature requirement serine protease A1 (HTRA1) in the pathogenesis of intervertebral disc (IVD) degeneration. J. Biol. Chem. 2012, 287, 21335–21345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.S.; Ahn, J.I. The effect of chondroitinase ABC on rabbit intervertebral disc—Radiological, histological and electron-microscopic finding. Int. Orthop. 1995, 19, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Creemers, L. Mechanisms of intervertebral disk degeneration/injury and pain: A review. Glob. Spine J. 2013, 3, 145–152. [Google Scholar]
- Kang, J.D.; Georgescu, H.I.; McIntyre-Larkin, L.; Stefanovic-Racic, M.; Donaldson, W.F., III; Evans, C.H. Herniated lumbar intervertebral discs spontaneously produce matrix metalloproteinases, nitric oxide, interleukin-6, and prostaglandin E2. Spine 1996, 21, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.D.; Stefanovic-Racic, M.; McIntyre, L.A.; Georgescu, H.I.; Evans, C.H. Toward a biochemical understanding of human intervertebral disc degeneration and herniation: Contributions of nitric oxide, interleukins, prostaglandin E2, and matrix metalloproteinases. Spine 1997, 22, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.M.; Doyle, P.; Minogue, B.M.; Gnanalingham, K.; Hoyland, J.A. Increased expression of matrix metalloproteinase-10, nerve growth factor and substance P in the painful degenerate intervertebral disc. Arthritis Res. Ther. 2009, 11, R126. [Google Scholar] [CrossRef] [PubMed]
- Purmessur, D.; Freemont, A.J.; Hoyland, J.A. Expression and regulation of neurotrophins in the nondegenerate and degenerate human intervertebral disc. Arthritis Res. Ther. 2008, 10, R99. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Evans, H.; Trivedi, J.; Menage, J. Histology and pathology of the human intervertebral disc. J. Bone Jt. Surg. 2006, 88 (Suppl. S2), 10–14. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.D.; Darby, S. Basic and clinical anatomy of the spine, spinal cord, and ANS. J. Manip. Physiol. Ther. 1997, 20, 294. [Google Scholar]
- Mulleman, D.; Mammou, S.; Griffoul, I.; Watier, H.; Goupille, P. Pathophysiology of disk-related sciatica. I.—Evidence supporting a chemical component. Jt. Bone Spine 2006, 73, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Ponnappan, R.K.; Markova, D.Z.; Antonio, P.J.D.; Murray, H.B.; Vaccaro, A.R.; Shapiro, I.M.; Anderson, D.G.; Albert, T.J.; Risbud, M.V. An organ culture system to model early degenerative changes of the intervertebral disc. Arthritis Res. Ther. 2011, 13, R171. [Google Scholar] [CrossRef] [PubMed]
- Sowa, G.A.; Coelho, J.P.; Vo, N.V.; Pacek, C.; Westrick, E.; Kang, J.D. Cells from degenerative intervertebral discs demonstrate unfavorable responses to mechanical and inflammatory stimuli: A pilot study. Am. J. Phys. Med. Rehabil. 2012, 91, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Hoogendoorn, R.J.; Wuisman, P.I.; Smit, T.H.; Everts, V.E.; Helder, M.N. Experimental intervertebral disc degeneration induced by chondroitinase ABC in the goat. Spine 2007, 32, 1816–1825. [Google Scholar] [CrossRef] [PubMed]
- Chiba, K.; Masuda, K.; Andersson, G.B.; Momohara, S.; Thonar, E.J. Matrix replenishment by intervertebral disc cells after chemonucleolysis in vitro with chondroitinase ABC and chymopapain. Spine J. 2007, 7, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Juliana, T.Y.; Lee, K.M.C.C.; Leung, V.Y.L. Extraction of rna from tough tissues with high proteoglycan content by cryosection, second phase separation and high salt precipitation. J. Biol. Methods 2015, 2, e20. [Google Scholar]
- Farndale, R.W.; Sayers, C.A.; Barrett, A.J. A direct spectrophotometric microassay for sulfated glycosaminoglycans in cartilage cultures. Connect. Tissue Res. 1982, 9, 247–248. [Google Scholar] [CrossRef] [PubMed]
- Dudli, S.; Haschtmann, D.; Ferguson, S.J. Persistent degenerative changes in the intervertebral disc after burst fracture in an in vitro model mimicking physiological post-traumatic conditions. Eur. Spine J. 2015, 24, 1901–1908. [Google Scholar] [CrossRef] [PubMed]

| Gene | Mean Cq ± SD | Gene | Mean Cq ± SD |
|---|---|---|---|
| S18 | 28.24 ± 5.22 | Collagen 2 | 26.92 ± 2.85 |
| GAPDH | 29.69 ± 1.78 | Aggrecan | 27.13 ± 3.33 |
| Actin-β | 30.02 ± 2.98 | MMP3 | 33.65 ± 3.55 |
| Collagen 1 | 35.52 ± 3.14 | TIMP1 | 27.78 ± 2.49 |
| Experimental Groups | Analyses | |
|---|---|---|
| Treatments | Gene Expression | ECM Composition |
| 1. Native tissue 2. Stable NP culture | n = 8 Day 3: n = 8 Day 7: n = 8 Day 14: n = 8 Day 21: n = 8 | n = 5 Day 3: n = 4 Day 7: n = 4 Day 14: n = 4 Day 21: n = 4 |
| Enzyme Treatment | ||
| 1. PBS 2. Chondroitinase ABC 1 U/mL 3. Chondroitinase ABC 5 U/mL 4. Chondroitinase ABC 10 U/mL 5. Chondroitinase ABC 20 U/mL | Day 3: n = 4/treatment Day 7: n = 4/treatment Day 14: n = 4/treatment | Day 3: n = 7/treatment Day 7: n = 5/treatment Day 14: n = 5/treatment |
| Cytokine Treatment | ||
| 1. Control 2. TNF-α + IL-1β (100 ng/mL) 3. EGCG (10 µM) 4. TNF-α + IL-1β + EGCG | Day 3: n = 5/treatment Day 7: n = 5/treatment Day 14: n = 7/treatment | Day 3: n = 4/treatment Day 7: n = 4/treatment Day 14: n = 4/treatment |
| Total | n = 321 NPs | |
| Gene | Primer Seq. No. | Gene | Primer Seq. No. |
|---|---|---|---|
| S18 | Bt03225193_m1 | ADAMTS4 | Bt03224693_m1 |
| GAPDH | Bt03210919_g1 | MMP1 | Bt04259497_m1 |
| Actin-β | Bt03279174_g1 | MMP3 | Bt04259495_m1 |
| Interleukin-6 | Bt03211904_m1 | MMP13 | Bt03214052_m1 |
| Interleukin-8 | Bt03211908_m1 | TIMP1 | Bt03223720_m1 |
| Interleukin-1β | Bt03212741_m1 | Collagen 1 | Bt03214883_m1 |
| iNOS | Bt03249584_m1 | Collagen 2 | Bt03251861_m1 |
| PTGS2 | Bt03214492_m1 | Aggrecan | Bt03212186_m1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krupkova, O.; Hlavna, M.; Amir Tahmasseb, J.; Zvick, J.; Kunz, D.; Ito, K.; Ferguson, S.J.; Wuertz-Kozak, K. An Inflammatory Nucleus Pulposus Tissue Culture Model to Test Molecular Regenerative Therapies: Validation with Epigallocatechin 3-Gallate. Int. J. Mol. Sci. 2016, 17, 1640. https://doi.org/10.3390/ijms17101640
Krupkova O, Hlavna M, Amir Tahmasseb J, Zvick J, Kunz D, Ito K, Ferguson SJ, Wuertz-Kozak K. An Inflammatory Nucleus Pulposus Tissue Culture Model to Test Molecular Regenerative Therapies: Validation with Epigallocatechin 3-Gallate. International Journal of Molecular Sciences. 2016; 17(10):1640. https://doi.org/10.3390/ijms17101640
Chicago/Turabian StyleKrupkova, Olga, Marian Hlavna, Julie Amir Tahmasseb, Joel Zvick, Dominik Kunz, Keita Ito, Stephen J. Ferguson, and Karin Wuertz-Kozak. 2016. "An Inflammatory Nucleus Pulposus Tissue Culture Model to Test Molecular Regenerative Therapies: Validation with Epigallocatechin 3-Gallate" International Journal of Molecular Sciences 17, no. 10: 1640. https://doi.org/10.3390/ijms17101640