Structure Conservation and Differential Expression of Farnesyl Diphosphate Synthase Genes in Euphorbiaceous Plants
Abstract
:1. Introduction
2. Results
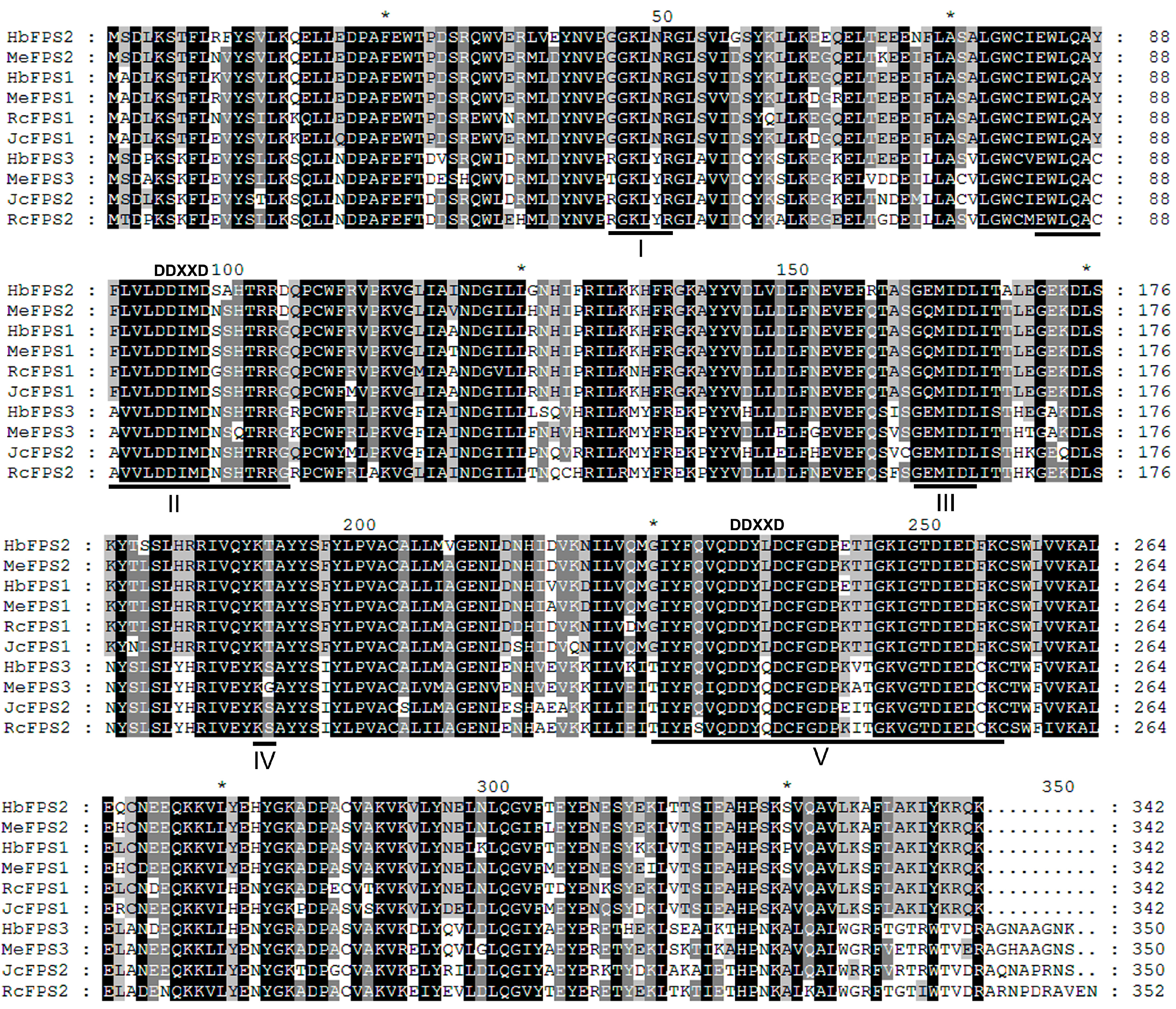
2.1. Cloning, Identification and Structure Analysis of the Euphorbiaceous Plants FPSs
| Gene | GenBank Accession No. | Gene Size (bp) | ORF (bp) | Predicted Protein | ||
|---|---|---|---|---|---|---|
| Size (aa) | MW (kDa) | pI | ||||
| HbFPS1 | Z49786 | 4690 | 1029 | 342 | 39.41 | 5.94 |
| HbFPS2 | KT306000 | 4171 | 1029 | 342 | 39.55 | 5.07 |
| HbFPS3 | KT306001 | 3710 | 1053 | 350 | 40.27 | 6.06 |
| MeFPS1 | KT306002 | 4349 | 1029 | 342 | 39.48 | 5.68 |
| MeFPS2 | KT306003 | 5666 | 1029 | 342 | 39.57 | 5.86 |
| MeFPS3 | KT306004 | 4296 | 1053 | 350 | 40.13 | 5.18 |
| RcFPS1 | KT306005 | 5720 | 1029 | 342 | 39.37 | 5.30 |
| RcFPS2 | XM_002522756 | 3583 | 1059 | 352 | 40.73 | 4.85 |
| JcFPS1 | XM_012219426 | 3977 | 1029 | 342 | 39.43 | 5.30 |
| JcFPS2 | XM_012215689 | 3886 | 1053 | 350 | 40.80 | 5.72 |
| HbFPS2 | HbFPS3 | MeFPS1 | MeFPS2 | MeFPS3 | RcFPS1 | RcFPS2 | JcFPS1 | JcFPS2 | EpFPS | |
|---|---|---|---|---|---|---|---|---|---|---|
| HbFPS1 | 90.94 | 68.71 | 95.61 | 94.44 | 67.84 | 91.81 | 69.30 | 93.27 | 66.37 | 89.47 |
| HbFPS2 | 65.20 | 90.64 | 91.81 | 65.20 | 86.84 | 66.08 | 87.43 | 63.45 | 84.50 | |
| HbFPS3 | 68.13 | 69.88 | 87.14 | 68.42 | 84.29 | 67.84 | 84.86 | 67.84 | ||
| MeFPS1 | 94.74 | 67.84 | 90.64 | 68.71 | 92.98 | 64.91 | 89.77 | |||
| MeFPS2 | 69.30 | 90.94 | 68.71 | 91.81 | 66.96 | 89.47 | ||||
| MeFPS3 | 66.96 | 81.71 | 66.08 | 84.00 | 66.08 | |||||
| RcFPS1 | 67.54 | 91.52 | 65.79 | 89.18 | ||||||
| RcFPS2 | 67.25 | 82.57 | 66.67 | |||||||
| JcFPS1 | 66.37 | 90.64 | ||||||||
| JcFPS2 | 64.91 |
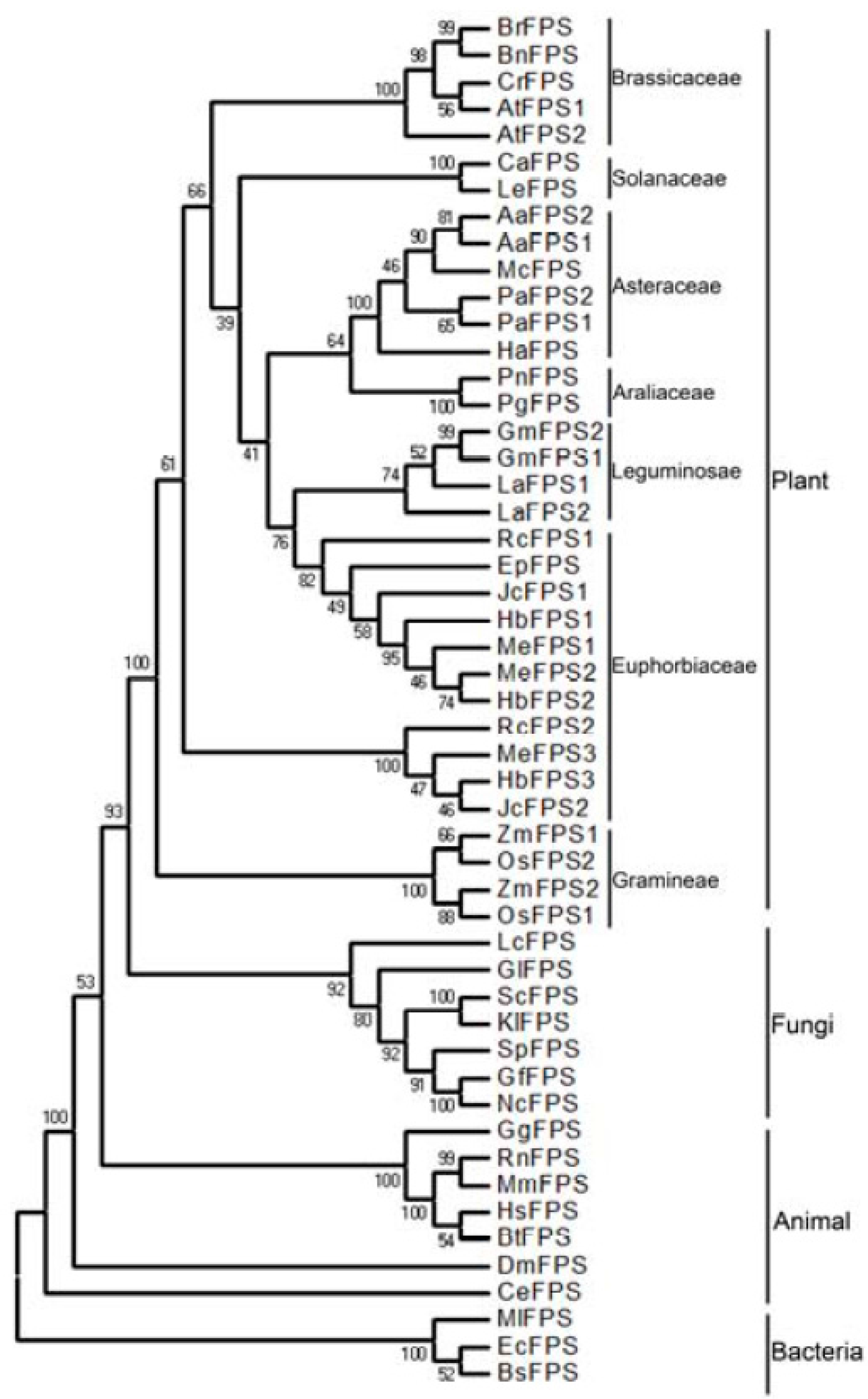
2.2. Phylogenetic Analysis
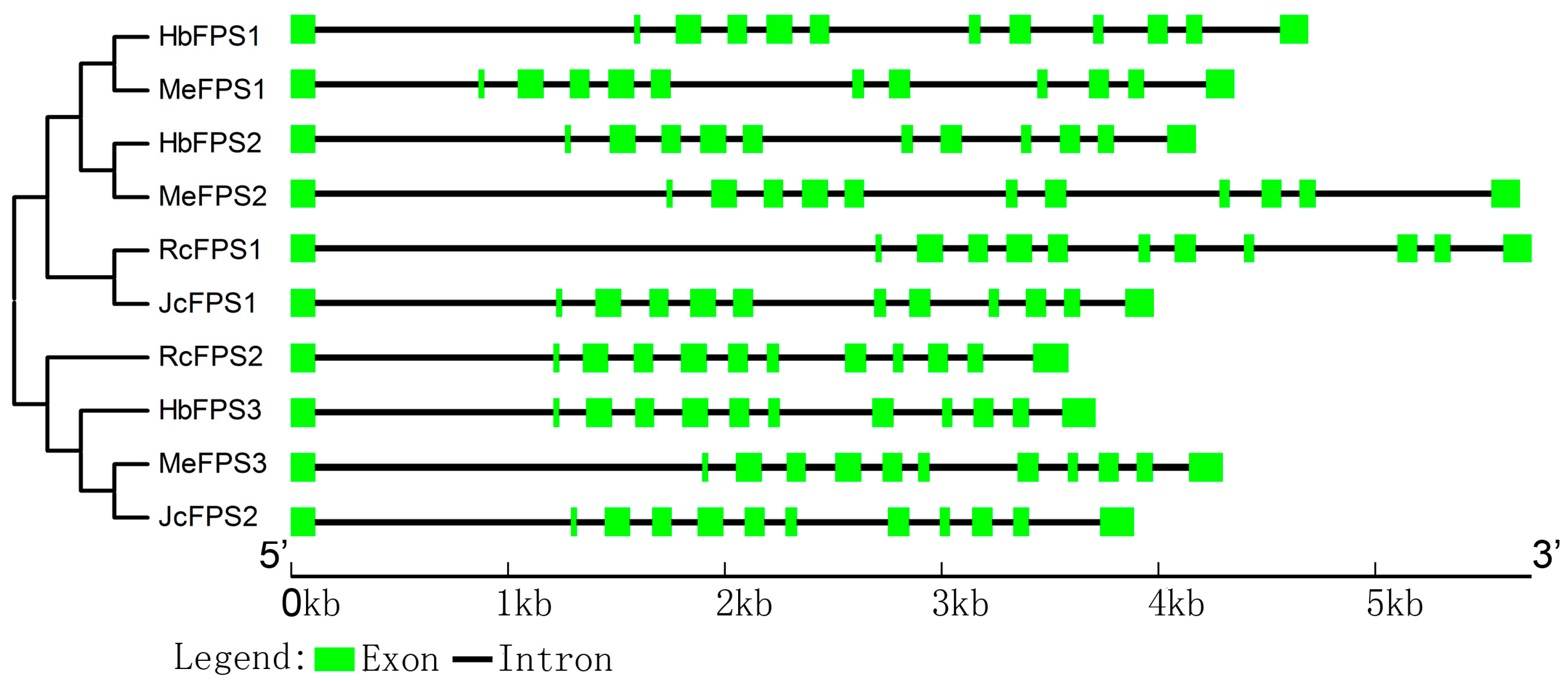
2.3. Intron and Exon Organization of FPSs
2.4. Structure Prediction and Homology Modeling of the FPSs
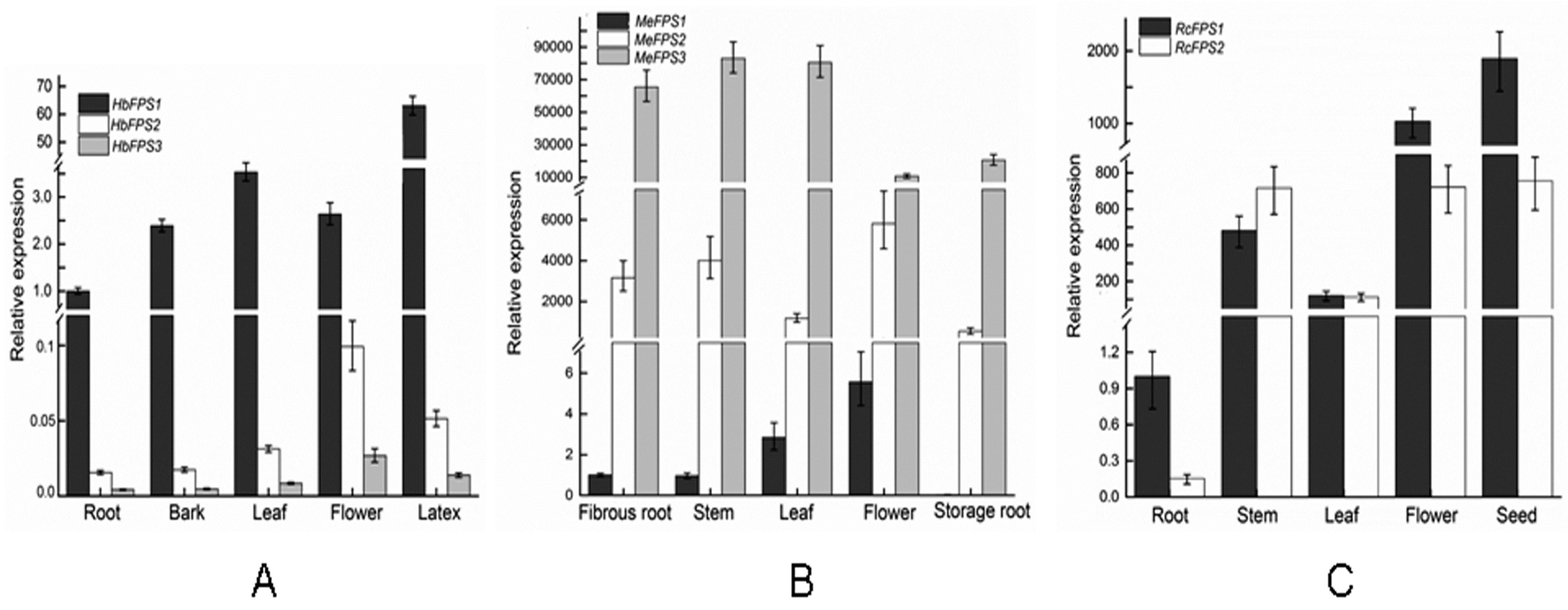
2.5. Expression Analysis of FPSs in Euphorbiaceous Plants Tissues

3. Discussion
4. Experimental Section
4.1. Plant Materials and Treatments
4.2. Cloning and Identification of FPS Genes
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| HbFPS1 | TCCATGGCGGATCTGAAGTCAACT | CATCCAGTCTTTGTCCATGTATCTG |
| HbFPS2 | AATCCATGTCTGATCTGAAGTCGA | ATCCAATCTTTGTCCATGTTCTTG |
| HbFPS3 | ATGAGCGATCCAAAATCCAAGTTCTTGG | ATGTTAATCCTCAGCTCATTTTAGAGT |
| MeFPS1 | CTCTGTTTTCAGTTTTTCTCCCCAATCT | CAATCTTTATCCATGTATCTGGATA |
| MeFPS2 | CACTCTTCATTCACTCG AATCTCCG | CATATTAAGTGTTTACTTAAATAATAA |
| MeFPS3 | GATATGAGCCAGTAAAGTTCCACAGTT | TTCTGAACCATTAGAAGAACAAGAAC |
| RcFPS1 | AGCTTCATTCATTCTTTTCTCTCC | GATGATAAAAACCATTCATTCAATT |
| RcFPS2 | GATTCAGAATTGTTCTTCAAAAGCGC | GAATCACAAAGTTGACAAGGAACCC |
| JcFPS1 | TCAATCTCTCCTCACTACTGCCCTCC | CGCATTATTCGGCATCATCCAATCAT |
| JcFPS2 | GCCCTTTCATATCGAACGGTAATAACAT | AAGTTTCATTTCCCATTCTAATGTTC |
4.3. Homology Modeling and Structure Prediction
4.4. Expression Analysis
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| HbFPS1 | TGAAAGCTATAAGAAACTAGTAACCTCT | TCATCCAGTCTTTGTCCATGTATC |
| HbBFPS2 | GAACGAAAGCTATGAGAAACTAACC | TCATCCAATCTTTGTCCATGTTCT |
| HbFPS3 | GGAACCAGATGGACAGTTGATAG | ACTAGGCAAATGCTGGTAATAGG |
| HbACT | CACCACCAGAGAGAAAGTACAG | GATGGACCAGACTCATCGTATTC |
| MeFPS1 | GAAAGCTATGAGATATTAGTGACT | ATCATCATCATTCAATCTTTATCCA |
| MeFPS2 | AAAGCTATGAGAAACTAGTAACCT | CCCTGTTTTTATTTATTTCTGTCT |
| MeFPS3 | AACCAGATGGACAGTTGAGAGAG | AAGAACAAGAACCAAAGCAGATG |
| MeACT | CAGTGGTCGACAACTGGTAT | ATCCTCCAATCCAGACACTGT |
| RcFPS1 | AGTGTTGAAGTCTTTCCTGGC | CTAGCATTATTCGCACGATCC |
| RcFPS2 | GCTTTGTGGGGAAGATTTACAG | ACAAAGTTGACAAGGAACCCAA |
| Rc18S RNA | TTGGTGGAGCGATTTGTC | CCCAGAACATCTAAGGGCAT |
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zeng, C.; Wang, W.; Zheng, Y.; Chen, X.; Bo, W.; Song, S.; Zhang, W.; Peng, M. Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res. 2009, 38, 3981–3995. [Google Scholar] [CrossRef] [PubMed]
- Archer, B.L.; Audley, B.G. New aspects of rubber biosynthesis. Bot. J. Linn. Soc. 1987, 94, 181–196. [Google Scholar] [CrossRef]
- Schmitz, P.M.; Kavallari, A. Crop plants versus energy plants—On the international food crisis. Bioorg. Med. Chem. 2009, 17, 4020–4021. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.P.; Crabtree, J.; Zhao, Q.; Lorenzi, H.; Orvis, J.; Puiu, D.; Melake-Berhan, A.; Jones, K.M.; Redman, J.; Chen, G.; et al. Draft genome sequence of the ricin-producing oilseed castor bean. Nat. Biotechnol. 2010, 28, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Wu, Y.; Banerjee, R.; Li, Y.; Yan, H.; Sharkey, T.D. Feedback inhibition of deoxy-d-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J. Biol. Chem. 2013, 288, 16926–16936. [Google Scholar] [CrossRef] [PubMed]
- Rasulov, B.; Talts, E.; Kännaste, A.; Niinemets, Ü. Bisphosphonate inhibitors reveal a large elasticity of plastidic isoprenoid synthesis pathway inisoprene-emitting hybrid aspen. Plant. Physiol. 2015, 168, 532–548. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Concepción, M. Early steps in isoprenoid biosynthesis: Multilevel regulation of the supply of common precursors in plant cells. Phytochem. Rev. 2006, 5, 1–15. [Google Scholar] [CrossRef]
- Lombard, J.; Moreira, D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 2011, 28, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007, 282, 21573–21577. [Google Scholar] [CrossRef] [PubMed]
- Eisenreich, W.; Bacher, A.; Arigoni, D.; Rohdich, F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell. Mol. Life Sci. 2004, 61, 1401–1426. [Google Scholar] [CrossRef] [PubMed]
- Bick, J.A.; Lange, B.M. Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: Unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 2003, 415, 146–154. [Google Scholar] [CrossRef]
- Chow, K.S.; Wan, K.L.; Mat, I.M.N.; Bahari, A.; Tan, S.H.; Harikrishna, K.; Yeang, H.Y. Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J. Exp. Bot. 2007, 58, 2429–2440. [Google Scholar] [CrossRef] [PubMed]
- Dhar, M.K.; Koul, A.; Kaul, S. Farnesyl pyrophosphate synthase: A key enzyme in isoprenoid biosynthetic pathway and potential molecular target for drug development. New Biotechnol. 2013, 30, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Sando, T.; Takaoka, C.; Mukai, Y.; Yamashita, A.; Hattori, M.; Ogasawara, N.; Fukusaki, E.; Kobayashi, A. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea Brasiliensis. Biosci. Biotechnol. Biochem. 2008, 72, 2049–2060. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Kroon, P.A.; Poulter, C.D. Isoprenyl diphosphate synthases: Protein sequence comparisons, a phylogenetic tree, and predictions of secondary structure. Protein Sci. 1994, 3, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009, 37, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Chen, X.; Zhang, M.; Su, P.; Liu, Y.J.; Tong, Y.R.; Wang, X.J.; Huang, L.Q.; Gao, W. Molecular cloning and characterisation of farnesyl pyrophosphate synthase from Tripterygium wilfordii. PLoS ONE 2015, 10, 125415. [Google Scholar] [CrossRef] [PubMed]
- Keim, V.; Manzano, D.; Fernández, F.J.; Closa, M.; Andrade, P.; Caudepón, D.; Bortolotti, C.; Vega, M.C.; Arró, M.; Ferrer, A. Characterization of Arabidopsis FPS isozymes and FPS gene expression analysis provide insight into the biosynthesis of isoprenoid precursors in seeds. PLoS ONE 2012, 7, 49109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunillera, N.; Arró, M.; Delourme, D.; Karst, F.; Boronat, A.; Ferrer, A. Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J. Biol. Chem. 1996, 271, 7774–7780. [Google Scholar] [PubMed]
- Cunillera, N.; Boronat, A.; Ferrer, A. The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphatesynthase isoform. J. Biol. Chem. 1997, 272, 15381–15388. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Cervantes, M.; Gallagher, C.E.; Zhu, C.; Wurtzel, E.T. Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant. Physiol. 2006, 141, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Hemmerlin, A.; Rivera, S.B.; Erickson, H.K.; Poulter, C.D. Enzymes encoded by the farnesyl diphosphate synthase gene family in the Big Sagebrush Artemisia tridentata ssp. spiciformis. J. Biol. Chem. 2003, 278, 32132–32140. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, M.G.; Lorenzi, P.; Sangalli, A.; Diani, E.; Mottes, M. Characterization and functional analysis of cis-acting elements of the human farnesyl diphosphate synthetase (FDPS) gene 5' flanking region. Genomics 2009, 93, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Adiwilaga, K.; Kush, A. Cloning and characterization of cDNA encoding farnesyl diphosphate synthase from rubber tree (Hevea brasiliensis). Plant Mol. Biol. 1996, 30, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Yin, T.; Miao, Q.; Li, C.; Ju, X.; Sun, Y.; Jiang, J. Molecular characterization and expression analysis of a gene encoding for farnesyl diphosphate synthase from Euphorbia pekinensis Rupr. Mol. Biol. Rep. 2012, 39, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Cunillera, N.; Boronat, A.; Ferrer, A. Spatial and temporal patterns of GUS expression directed by 59 regions of the Arabidopsis thaliana farnesyl diphosphate synthase genes FPS1 and FPS2. Plant Mol. Biol. 2000, 44, 745–758. [Google Scholar] [CrossRef]
- Closa, M.; Vranová, E.; Bortolotti, C.; Bigler, L.; Arró, M.; Ferrer, A.; Gruissem, W. The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J. 2010, 63, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liao, Z.; Guo, L.; Li, W.; Chen, M.; Pi, Y.; Gong, Y.; Sun, X.; Tang, K. Cloning and functional analysis of a cDNA encoding Ginkgo biloba farnesyl diphosphate yynthase. Mol. Cells 2004, 18, 150–156. [Google Scholar] [PubMed]
- Carrier, D.J.; van Beek, T.A.; van der Heijden, R.; Verpoort, R. Distribution of ginkgolides and terpenoid biosynthetic activity in G. biloba. Phytochemistry 1998, 48, 89–92. [Google Scholar] [CrossRef]
- Tang, C.; Qi, J.; Li, H.; Zhang, C.; Wang, Y. A convenient and efficient protocol for isolating high quality RNA from latex of Hevea brasiliensis (para rubber tree). J. Biochem. Biophys. Methods 2007, 70, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.H.; Luo, M.W.; Zhang, C.F.; Zeng, H.C. Extraction of high-quality RNA from rubber tree leaves. Biosci. Biotechnol. Biochem. 2012, 76, 1394–1396. [Google Scholar] [CrossRef] [PubMed]
- Phytozome. Available online: http://www.phytozome.net/cassava (accessed on 15 April 2011).
- Jatropha Genome Database. Available online: http://www.kazusa.or.jp/jatropha (accessed on 1 February 2011).
- SWISS-MODEL. Available online: http://swissmodel.expasy.org (accessed on 20 January 2014).
- Li, H.L.; Guo, D.; Yang, Z.P.; Tang, X.; Peng, S.Q. Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 2014, 104, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Li, H.L.; Tang, X.; Peng, S.Q. Cassava (Manihot esculenta Krantz) genome harbors KNOX genes differentially expressed during storage root development. Genet. Mol. Res. 2014, 13, 10714–10726. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Zhang, L.B.; Guo, D.; Li, C.Z.; Peng, S.Q. Identification and expression profiles of the WRKY transcription factor family in Ricinus communis. Gene 2012, 503, 248–253. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, D.; Li, H.-L.; Peng, S.-Q. Structure Conservation and Differential Expression of Farnesyl Diphosphate Synthase Genes in Euphorbiaceous Plants. Int. J. Mol. Sci. 2015, 16, 22402-22414. https://doi.org/10.3390/ijms160922402
Guo D, Li H-L, Peng S-Q. Structure Conservation and Differential Expression of Farnesyl Diphosphate Synthase Genes in Euphorbiaceous Plants. International Journal of Molecular Sciences. 2015; 16(9):22402-22414. https://doi.org/10.3390/ijms160922402
Chicago/Turabian StyleGuo, Dong, Hui-Liang Li, and Shi-Qing Peng. 2015. "Structure Conservation and Differential Expression of Farnesyl Diphosphate Synthase Genes in Euphorbiaceous Plants" International Journal of Molecular Sciences 16, no. 9: 22402-22414. https://doi.org/10.3390/ijms160922402





