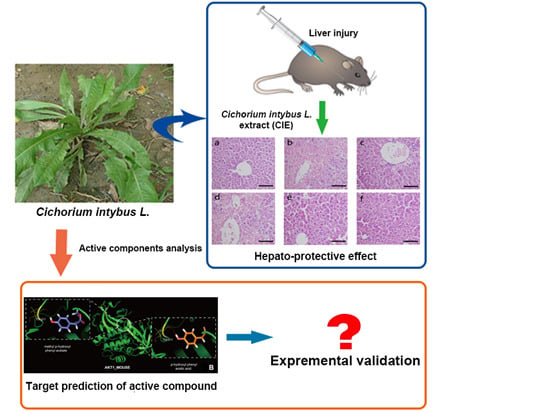
In Silico Analysis and Experimental Validation of Active Compounds from Cichorium intybus L. Ameliorating Liver Injury
Abstract
:1. Introduction
2. Results
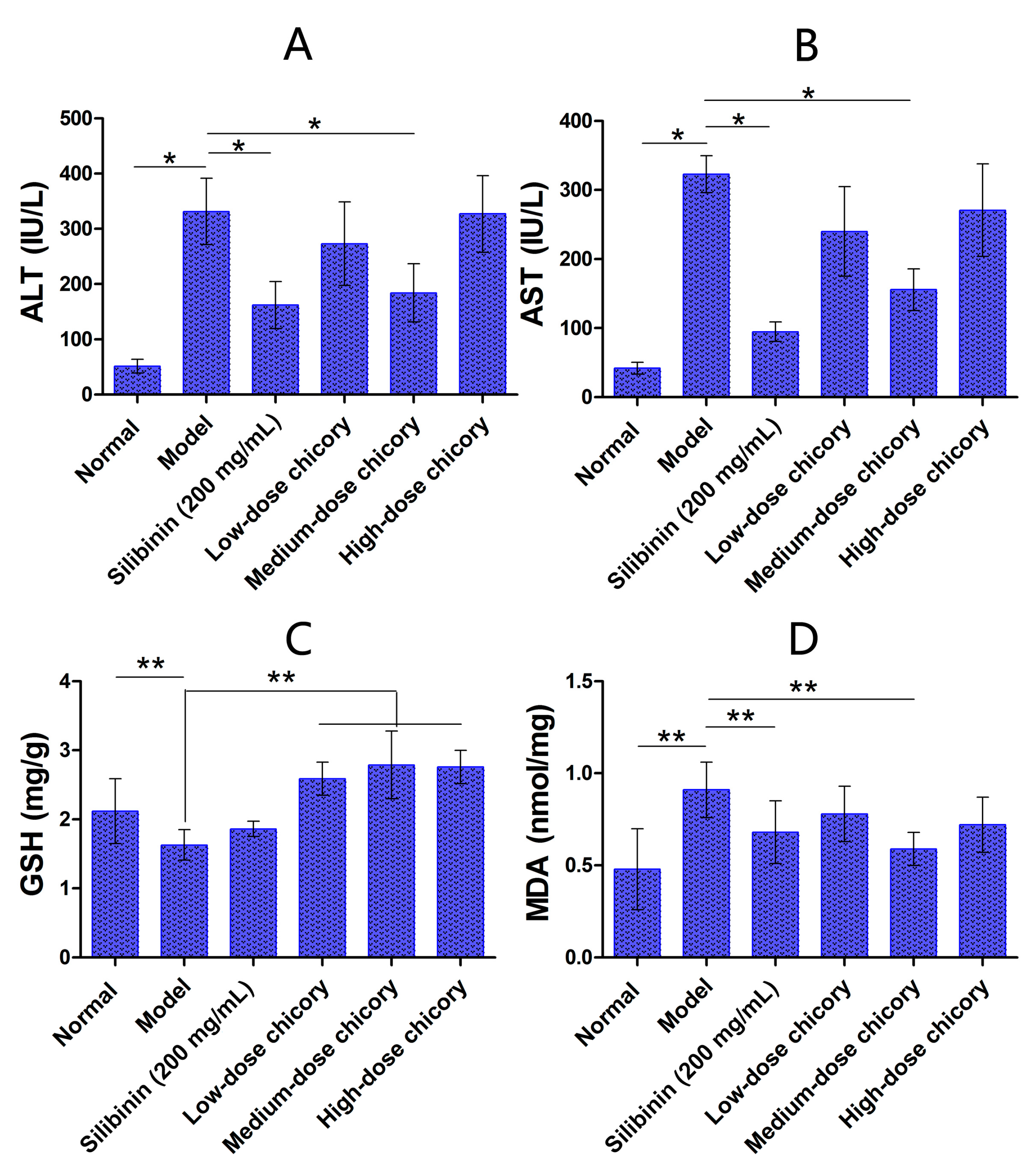
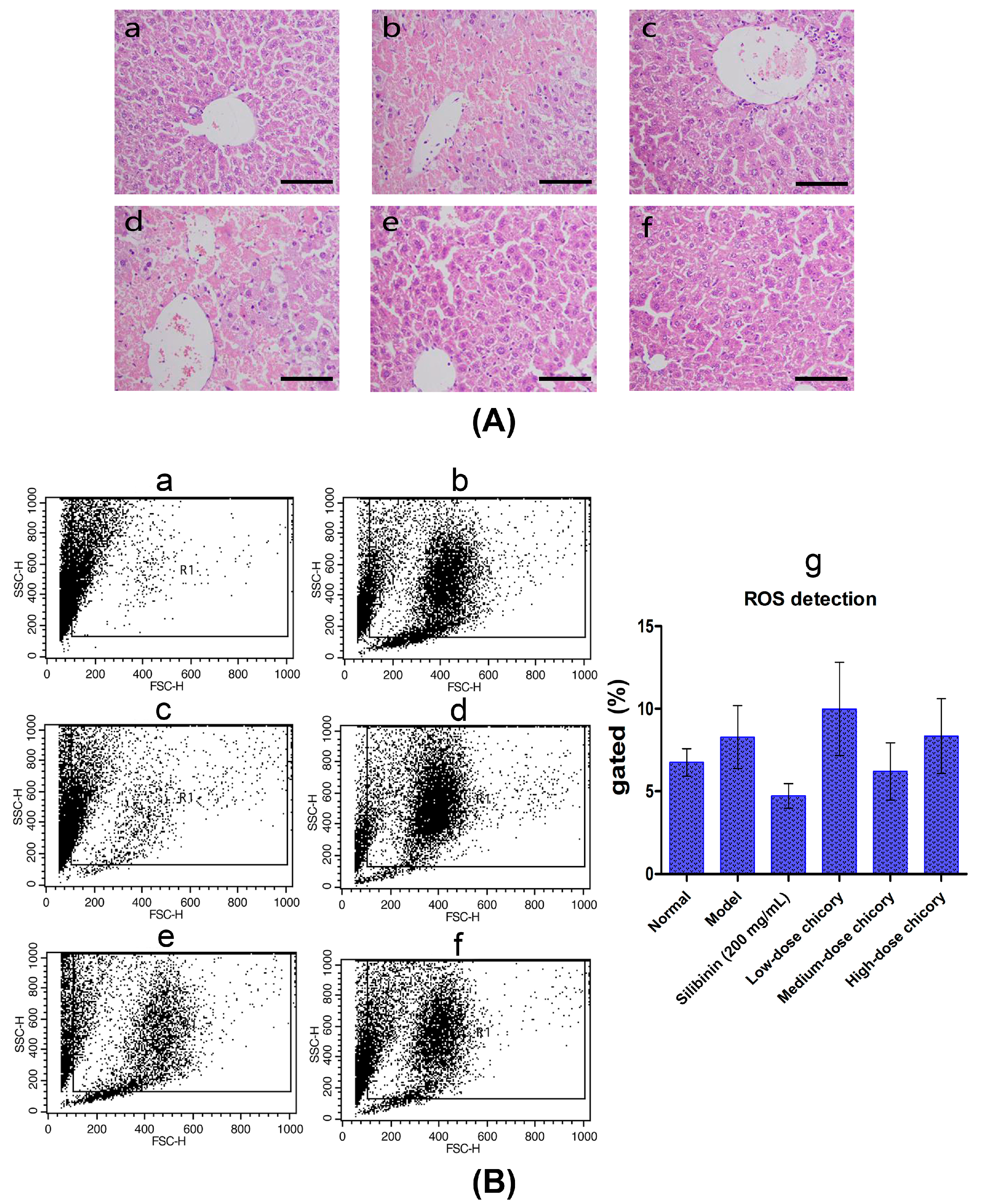
2.1. The Effect of Cichorium intybus L. on Liver Injury Protection
2.2. Small Molecule Compounds and Target Proteins
| Number | Name | Structure | Identification Means |
|---|---|---|---|
| 2 | Intyboate B |  | 13C-NMR; 1H-NMR; HMBC; HSQC; NOESY; m.p.; UV; CD; HR-ESI-TOF-MS |
| 5 | 3aR-santamarine |  | 13C-NMR; 1H-NMR; HMBC; HMQC; NOSEY; m.p.; UV; CD; HR-ESI-TOF-MS |
| 6 | Aurantiamide acetate |  | 13C-NMR; 1H-NMR; HMBC; HMQC; m.p.; UV; CD; HR-ESI-TOF-MS |
| 7 | Luteolin-7-O-β-d-glucoside |  | 13C-NMR; 1H-NMR; m.p.; HR-ESI-TOF-MS |
| 8 | Luteolin |  | 13C-NMR; 1H-NMR; HR-ESI-TOF-MS |
| 9 | Caffeic acid |  | 13C-NMR; 1H-NMR; HR-ESI-TOF-MS |
| 10 | Methyl 4-hydroxyphenylacetate |  | 13C-NMR; 1H-NMR; HMBC; HMQC; HR-ESI-TOF-MS |
| 11 | β-sitosterol |  | TLC |
| 12 | Daucosterol |  | TLC |
| 13 | 4-Hydroxylphenylacetic acid |  | HR-ESI-TOF-MS |
| Number | Name | ZINC | Target | UniProtKB AC | Recommended Name |
|---|---|---|---|---|---|
| 1 | Intyboate A | None | ACE_MOUSE_100 | P09470 | Angiotensin-converting enzyme |
| 2 | Intyboate B | None | ACE_MOUSE_100 | P09470 | Angiotensin-converting enzyme |
| 5 | 3aR-santamarine | ZINC05663288 | None | None | None |
| 7 | Luteolin-7-O-â-d-glucoside | ZINC03947434 | TOP1_MOUSE_10000 | Q04750 | DNA topoisomerase 1; Interleukin-5; Aldo-ketoreductase family 1 member C21; Multidrug resistance protein 3; γ-Aminobutyric acid receptor subunit α-1 |
| IL5_MOUSE_10000 | P04401 | ||||
| AK1CL_MOUSE_10000 | Q91WR5 | ||||
| MDR3_MOUSE_10000 | P21440 | ||||
| GBRA1_MOUSE_10 | P62812 | ||||
| 8 | Luteolin | ZINC18185774 | AK1CL_MOUSE_10000 | Q91WR5 | Aldo-ketoreductase family 1 member C21; Multidrug resistance protein 3; Tyrosinase; γ-Aminobutyric acid receptor subunit α-1; Interleukin-5; Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit α isoform; Dual specificity protein phosphatase 1; Solute carrier family 22 member 6 |
| MDR3_MOUSE_100 | P21440 | ||||
| TYRO_MOUSE_10000 | P11344 | ||||
| GBRA1_MOUSE_1000 | P62812 | ||||
| IL5_MOUSE_10000 | P04401 | ||||
| PK3CA_MOUSE_10000 | P42337 | ||||
| DUS1_MOUSE_10000 | P28563 | ||||
| S22A6_MOUSE_10000 | Q8VC69 | ||||
| 9 | Caffeic acid | ZINC00058172 | MDR2_MOUSE_10000 | None | Multidrug resistance protein 1B; Multidrug resistance protein 3; Transcription factor HES-1; Dual specificity tyrosine-phosphorylation-regulated kinase 1A; Epidermal growth factor receptor; Carbonic anhydrase 15; Retinoic acid receptor RXR-α |
| MDR1_MOUSE_10000 | P06795 | ||||
| MDR3_MOUSE_10000 | P21440 | ||||
| HES1_MOUSE_10000 | P35428 | ||||
| DYR1A_MOUSE_10000 | Q61214 | ||||
| EGFR_MOUSE_10000 | Q01279 | ||||
| CAH15_MOUSE_10000 | Q99N23 | ||||
| RXRA_MOUSE_1000 | P28700 | ||||
| 10 | Methyl 4-hydroxyphenylacetate | ZINC00395674 | ACE_MOUSE_1000 | P09470 | Angiotensin-converting enzyme; Solute carrier family 22 member 20; Solute carrier family 22 member 6; Platelet-derived growth factor receptor α (PDGFRα); Carbonic anhydrase 15; Mu-type opioid receptor; Bifunctional epoxide hydrolase 2; RAC-α serine/threonine-protein kinase (Akt-1); M-phase inducer phosphatase 2 |
| S22AK_MOUSE_10000 | Q80UJ1 | ||||
| S22A6_MOUSE_10000 | Q8VC69 | ||||
| PGFRA_MOUSE_1000 | P26618 | ||||
| CAH15_MOUSE_10000 | Q99N23 | ||||
| OPRM_MOUSE_10 | P42866 | ||||
| HYES_MOUSE_10000 | P34914 | ||||
| AKT1_MOUSE_10000 | P31750 | ||||
| MPIP2_MOUSE_10000 | P30306 | ||||
| 13 | 4-Hydroxylphenylacetic acid | ZINC00213065 | ACE_MOUSE_1000 | P09470 | Angiotensin-converting enzyme; Solute carrier family 22 member 6; Caspase-1; Glycine receptor subunit α-4; Solute carrier family 22 member 20; RAC-α serine/threonine-protein kinase (Akt-1); Prostaglandin G/H synthase 2; Carbonic anhydrase 15; Hydroxycarboxylic acid receptor 2; Platelet-derived growth factor receptor α(PDGFRα) |
| S22A6_MOUSE_10000 | Q8VC69 | ||||
| CASP1_MOUSE_10 | P29452 | ||||
| GLRA4_MOUSE_10000 | Q61603 | ||||
| S22AK_MOUSE_10000 | Q80UJ1 | ||||
| AKT1_MOUSE_10000 | P31750 | ||||
| MDR2_MOUSE_10000 | None | ||||
| PGH2_MOUSE_1000 | Q05769 | ||||
| CAH15_MOUSE_10000 | Q99N23 | ||||
| HCAR2_MOUSE_10000 | Q9EP66 | ||||
| PGFRA_MOUSE_1000 | P26618 |
2.3. Molecular Modeling, Molecular Docking and MD Simulation
2.4. The Effect of Compound 10 and Compound 13 on Disease Protection in Mice Hepatocytes
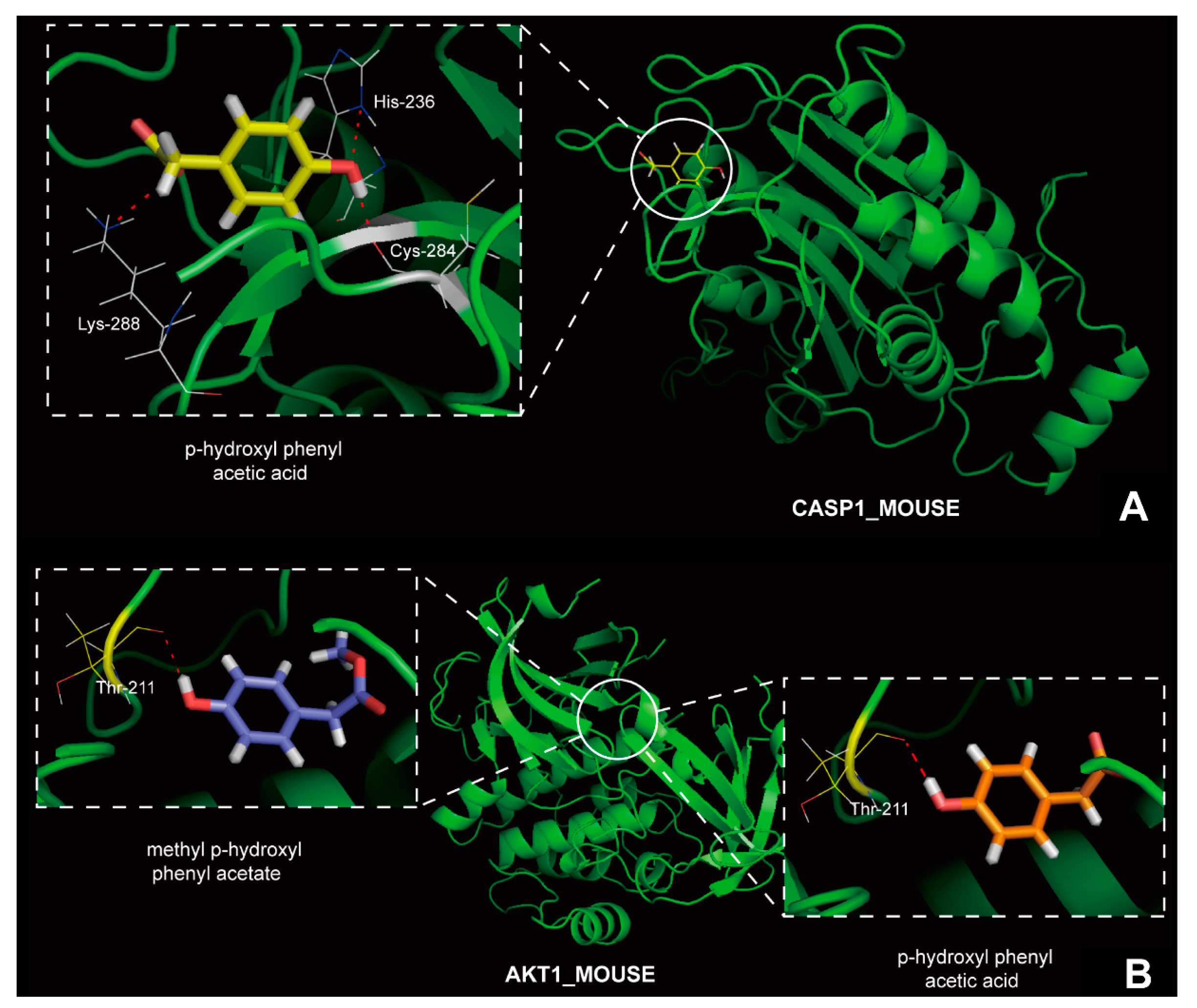

3. Discussion
4. Experimental Section
4.1. Experimental Animals and Design
4.2. Measurement of Markers of Liver Function and Enzyme Activity
4.3. Histopathological Observations in the Liver
4.4. Detection of ROS and Immunostaining
4.5. Isolation and Identification of Main Components in Cichorium intybus L.
4.6. Predictive Targets of Small Molecule Compounds
4.7. Molecular Modeling and Molecular Docking
4.8. MD Simulations of Complexes
4.9. Western Blot Analysis
4.10. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Hassan, H.A.; Yousef, M.I. Ameliorating effect of chicory (Cichorium intybus L.)-supplemented diet against nitrosamine precursors-induced liver injury and oxidative stress in male rats. Food Chem. Toxicol. 2010, 48, 2163–2169. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Y.; Gao, H.Y.; Huang, J.; Lu, J.; Gu, J.K.; Wang, J.H. Hepatoprotective effect of Cichorium intybus L., a traditional Uighur medicine, against carbon tetrachloride-induced hepatic fibrosis in rats. World J. Gastroenterol. 2014, 16, 4753–4760. [Google Scholar] [CrossRef] [PubMed]
- Saggu, S.; Sakeran, M.I.; Zidan, N.; Tousson, E.; Mohan, A.; Rehman, H. Ameliorating effect of chicory (Chichorium intybus L.) fruit extract against 4-tert-octylphenol induced liver injury and oxidative stress in male rats. Food Chem. Toxicol. 2014, 72, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.K.; Klausen, C.L.; Ekstrand, B. Regulation of cytochrome P450 mRNA expression in primary porcine hepatocytes by selected secondary plant metabolites from chicory (Cichorium intybus L.). Food Chem. 2014, 146, 255–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.Y.; Zhang, B.; Lin, Z.J.; Li, L.Y.; Wang, H.P.; Zhou, J. Chicory extract’s influence on gut bacteria of abdominal obesity rat. China J. Chin. Mater. Med. 2014, 11, 2081–2085. (In Chinese) [Google Scholar]
- Liu, H.Y.; Ivarsson, E.; Lundh, T.; Lindberg, J.E. Chicory (Cichorium intybus L.) and cereals differently affect gut development in broiler chickens and young pigs. J. Anim. Sci. Biotechnol. 2013, 1, 50. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, Y.S.; Lebda, M.A.; Hassinin, M.; Neoman, S.A. Chicory (Cichorium intybus L.) root extract regulates the oxidative status and antioxidant gene transcripts in CCl4-induced hepatotoxicity. PLoS ONE 2015, 10, e0121549. [Google Scholar] [CrossRef] [PubMed]
- Darr, J.; Klochendler, A.; Isaac, S.; Eden, A. Loss of IGFBP7 expression and persistent Akt activation contribute to SMARCB1/Snf5-mediated tumorigenesis. Oncogene 2014, 33, 3024–3032. [Google Scholar] [CrossRef] [PubMed]
- Edinger, A.L.; Thompson, C.B. Death by design: Apoptosis, necrosis and autophagy. Curr. Opin. Cell. Biol. 2004, 16, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. The expanding role of mitochondria in apoptosis. Genes. Dev. 2001, 15, 2922–2933. [Google Scholar] [PubMed]
- Hsieh, C.Y.; Chen, C.L.; Yang, K.C.; Ma, C.T.; Choi, P.C.; Lin, C.F. Detection of reactive oxygen species during the cell cycle under normal culture conditions using a modified fixed-sample staining method. J. Immunoass. Immunochem. 2015, 2, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.J.; Hsieh, C.Y.; Chen, C.L.; Yang, K.C.; Ma, C.T.; Choi, P.C.; Lin, C.F. A modified fixed staining method for the simultaneous measurement of reactive oxygen species and oxidative responses. Biochem. Biophys. Res. Commun. 2013, 1, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Macchia, I.; Urbani, F.; Proietti, E. Immune monitoring in cancer vaccine clinical trials: Critical issues of functional flow cytometry-based assays. Biomed. Res. Int. 2013, 2013, 726239. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.Y.; Lu, D.W.; Yeh, M.K.; Chiang, C.H. A novel high-content flow cytometric method for assessing the viability and damage of rat retinal ganglion cells. PLoS ONE 2012, 3, e33983. [Google Scholar] [CrossRef] [PubMed]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014, 7, 1126–1167. [Google Scholar] [CrossRef] [PubMed]
- Kühner, D.; Stahl, M.; Demircioglu, D.D.; Bertsche, U. From cells to muropeptide structures in 24 h: Peptidoglycan mapping by UPLC-MS. Sci. Rep. 2014, 4, 7494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Xu, X.F.; Huang, Y.; Wamg, D.Q.; Deng, Q.H. Characteristic fingerprint analysis and determination of main components on Andrographis paniculata extract by UPLC. J. Chin. Med. Mater. 2014, 6, 1055–1058. (In Chinese) [Google Scholar]
- Zhou, S.S.; Xu, J.D.; Shen, H.; Liu, H.H.; Li, S.L. Development of UPLC-Q-TOF-MS/MS combined with reference herb approach to rapidly screen commercial sulfur-fumigated ginseng. China J. Chin. Mater. Med. 2014, 15, 2807–2813. (In Chinese) [Google Scholar]
- Wojdyło, A.; Nowicka, P.; Laskowski, P.; Oszmiański, J. Evaluation of sour cherry (Prunus cerasus L.) fruits for their polyphenol content, antioxidant properties, and nutritional components. J. Agric. Food Chem. 2014, 51, 12332–12345. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y. High-Performance Liquid Chromatography (HPLC): Principles, Procedures and Practices; Nova Science Publishers: New York, NY, USA, 2014. [Google Scholar]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Wood, V.; Mungall, C.J.; Lewis, S.E.; Blake, J.A. On the use of gene ontology annotations to assess functional similarity among orthologs and paralogs: A short report. PLoS Comput. Biol. 2012, 2, e1002386. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Lang, P.T.; Brozell, S.R.; Mukherjee, S.; Pettersen, E.F.; Meng, E.C.; Thomas, V.; Rizzo, R.C.; Case, D.A.; James, T.L.; Kuntz, I.D. DOCK 6: Combining techniques to model RNA-small molecule complexes. RNA 2009, 15, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecularsimulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Liu, J.; Chen, Y.; Wang, F.T.; Wen, X.; Liu, H.Q.; Wang, M.Y.; Ouyang, L.; Huang, J.; Bao, J.K.; et al. In silico analysis and experimental validation of azelastine hydrochloride (N4) targetingsodium taurocholate co-transporting polypeptide (NTCP) in HBV therapy. Cell Prolif. 2014, 47, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzer, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Chen, X.; Guo, J.; Xi, R.X.; Chang, Y.W.; Pan, F.Y.; Zhang, X.Z. MiR-210 expression reverses radioresistance of stem-like cells of oesophageal squamous cell carcinoma. World J. Clin. Oncol. 2014, 5, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Song, J.; Park, C. Determining protein stability in cell lysates by pulse proteolysis and western blotting. Protein Sci. 2009, 5, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Bai, H.; Wang, X.Y.; Li, L.Y.; Cao, Y.H.; Wei, J.; Liu, Y.M.; Liu, L.J.; Gong, X.D.; Wu, L.; et al. Identification and validation of rice reference proteins for western blotting. J. Exp. Bot. 2011, 14, 4763–4772. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.-Y.; Zheng, Y.-X.; Sun, F.-Z.; Huang, J.; Lou, M.-M.; Gu, J.-K.; Wang, J.-H. In Silico Analysis and Experimental Validation of Active Compounds from Cichorium intybus L. Ameliorating Liver Injury. Int. J. Mol. Sci. 2015, 16, 22190-22204. https://doi.org/10.3390/ijms160922190
Li G-Y, Zheng Y-X, Sun F-Z, Huang J, Lou M-M, Gu J-K, Wang J-H. In Silico Analysis and Experimental Validation of Active Compounds from Cichorium intybus L. Ameliorating Liver Injury. International Journal of Molecular Sciences. 2015; 16(9):22190-22204. https://doi.org/10.3390/ijms160922190
Chicago/Turabian StyleLi, Guo-Yu, Ya-Xin Zheng, Fu-Zhou Sun, Jian Huang, Meng-Meng Lou, Jing-Kai Gu, and Jin-Hui Wang. 2015. "In Silico Analysis and Experimental Validation of Active Compounds from Cichorium intybus L. Ameliorating Liver Injury" International Journal of Molecular Sciences 16, no. 9: 22190-22204. https://doi.org/10.3390/ijms160922190