Genome-Wide Identification and Expression Analyses of Aquaporin Gene Family during Development and Abiotic Stress in Banana
Abstract
:1. Introduction
2. Results
2.1. Identification of AQP Genes in Banana
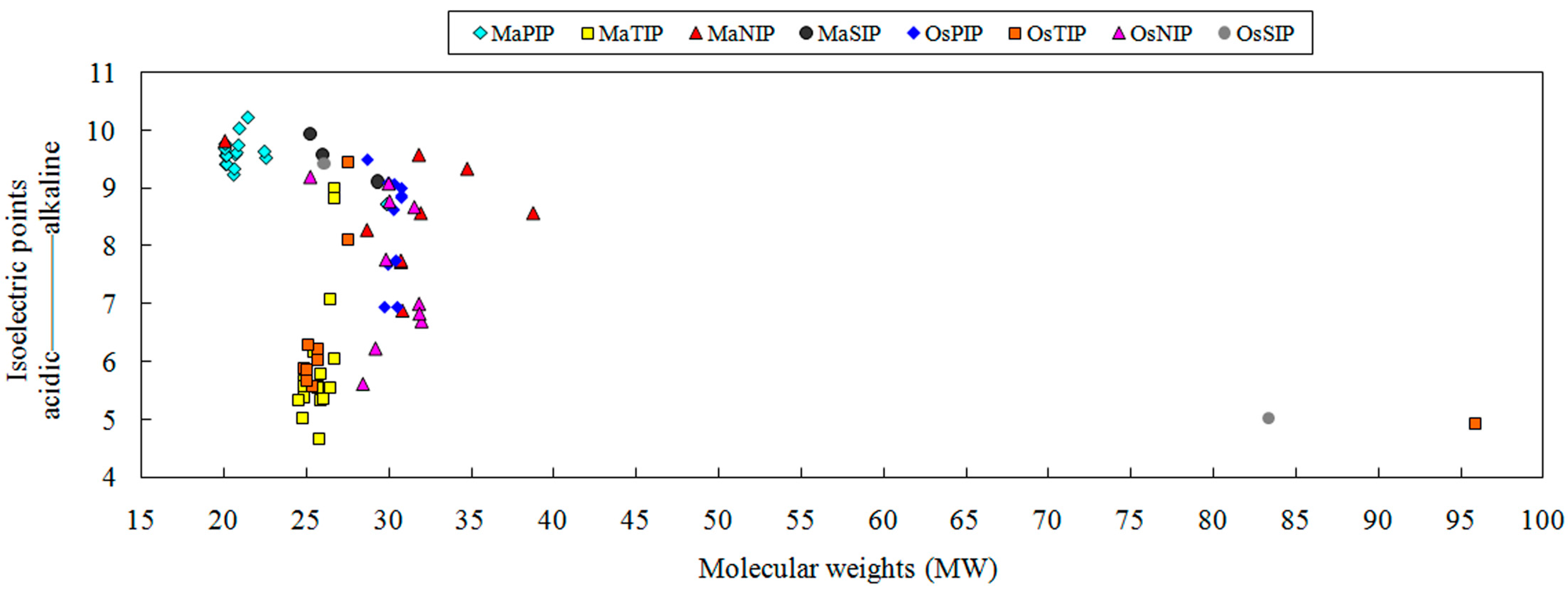
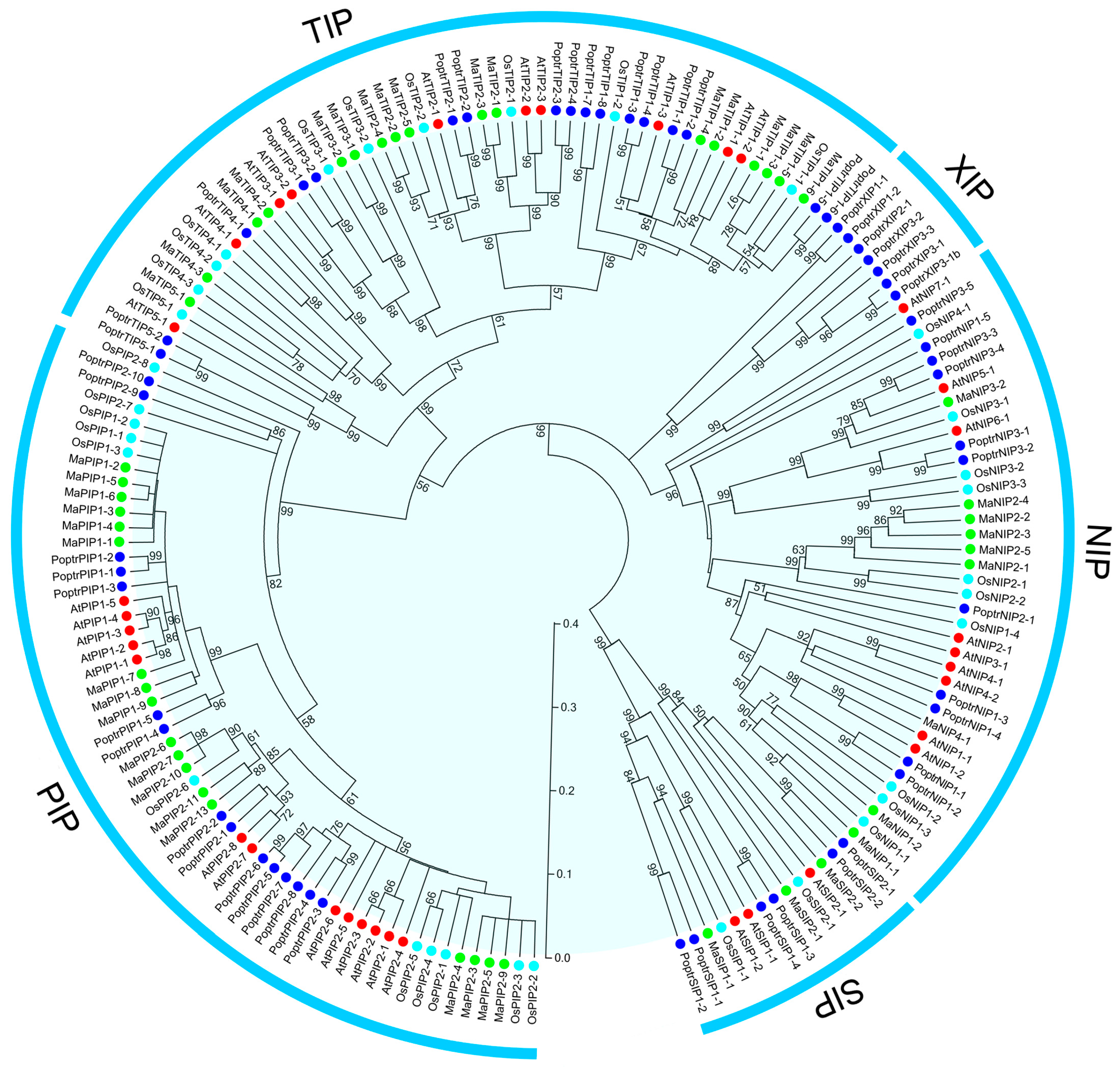
2.2. Phylogenetic Analysis of AQP Proteins
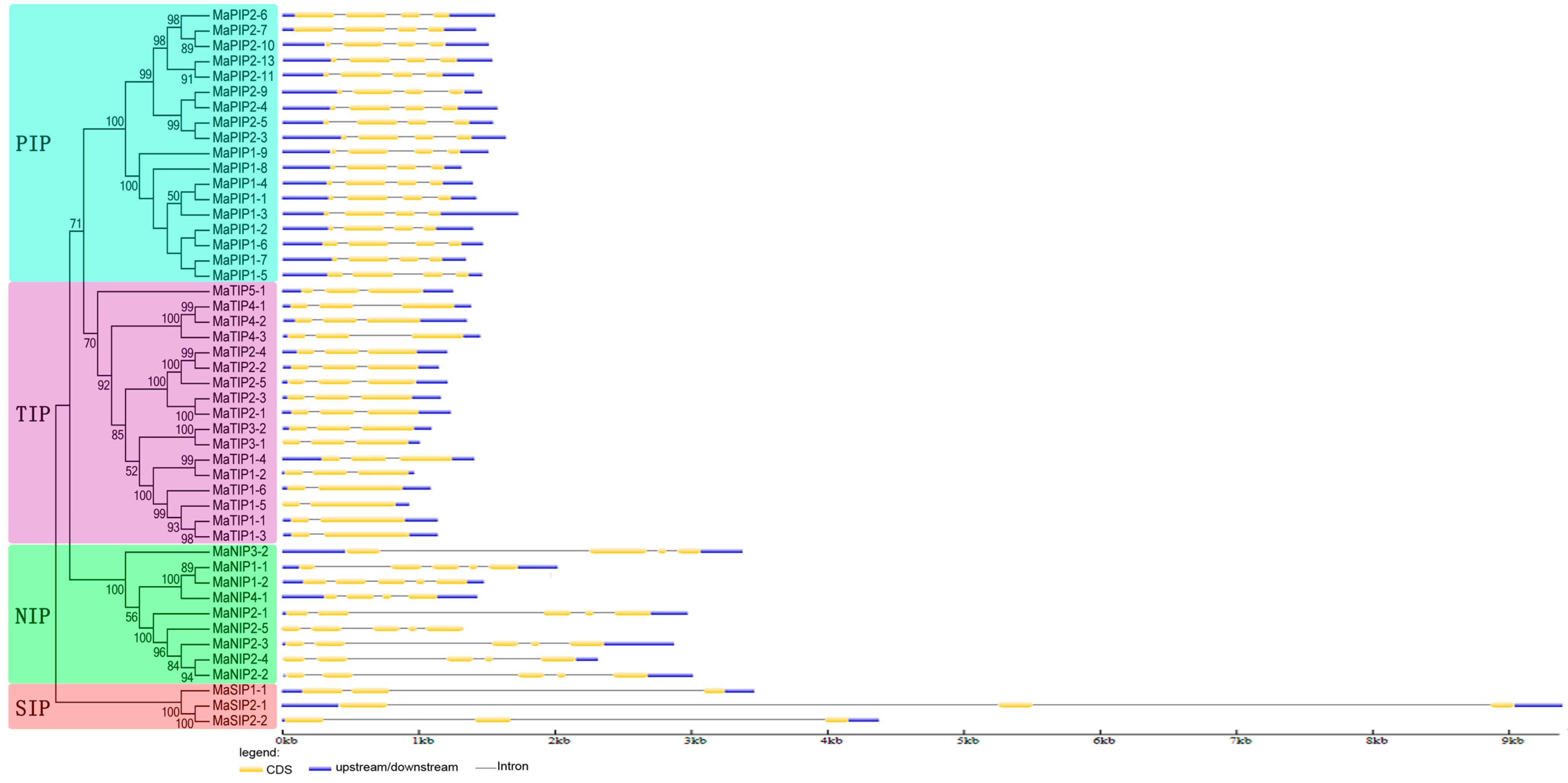
2.3. Gene Structure of AQP Genes in Banana
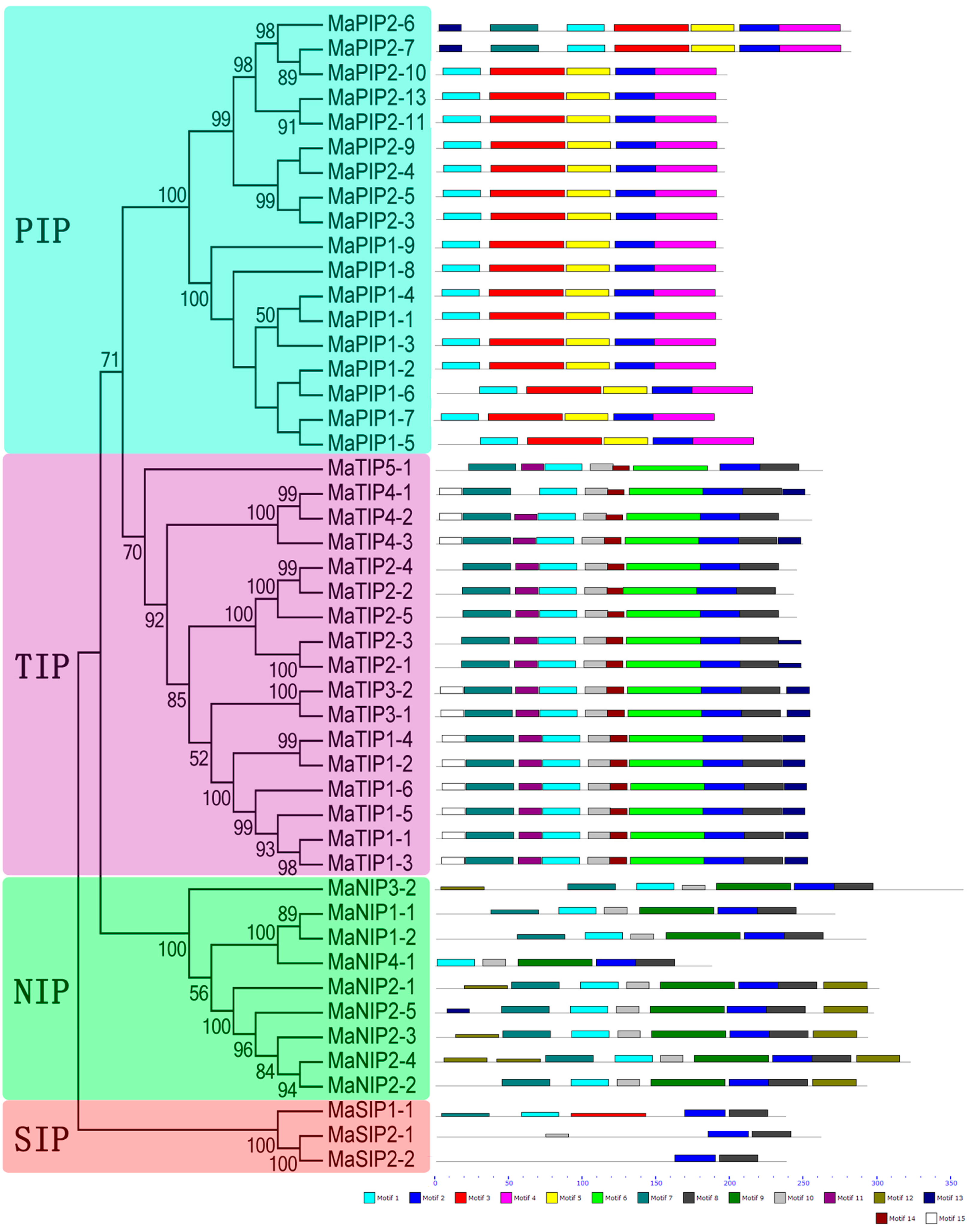
2.4. Conserved Motifs of AQP Proteins in Banana
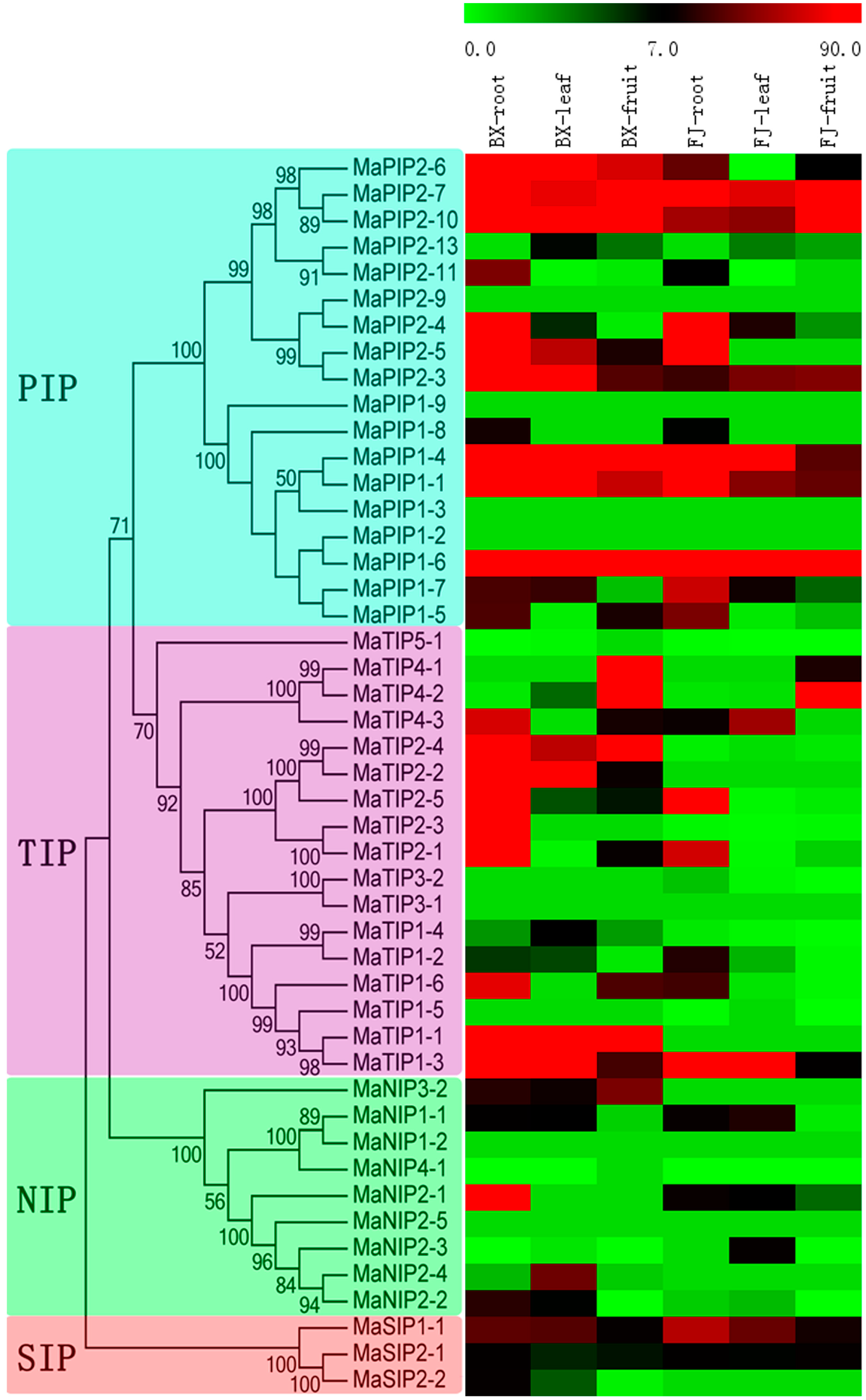
2.5. Expression Analysis of MaAQP Genes in Different Tissues of Two Banana Varieties
2.6. Expression Analysis of MaAQP Genes in Different Stages of Fruit Development and Ripening of Two Banana Varieties
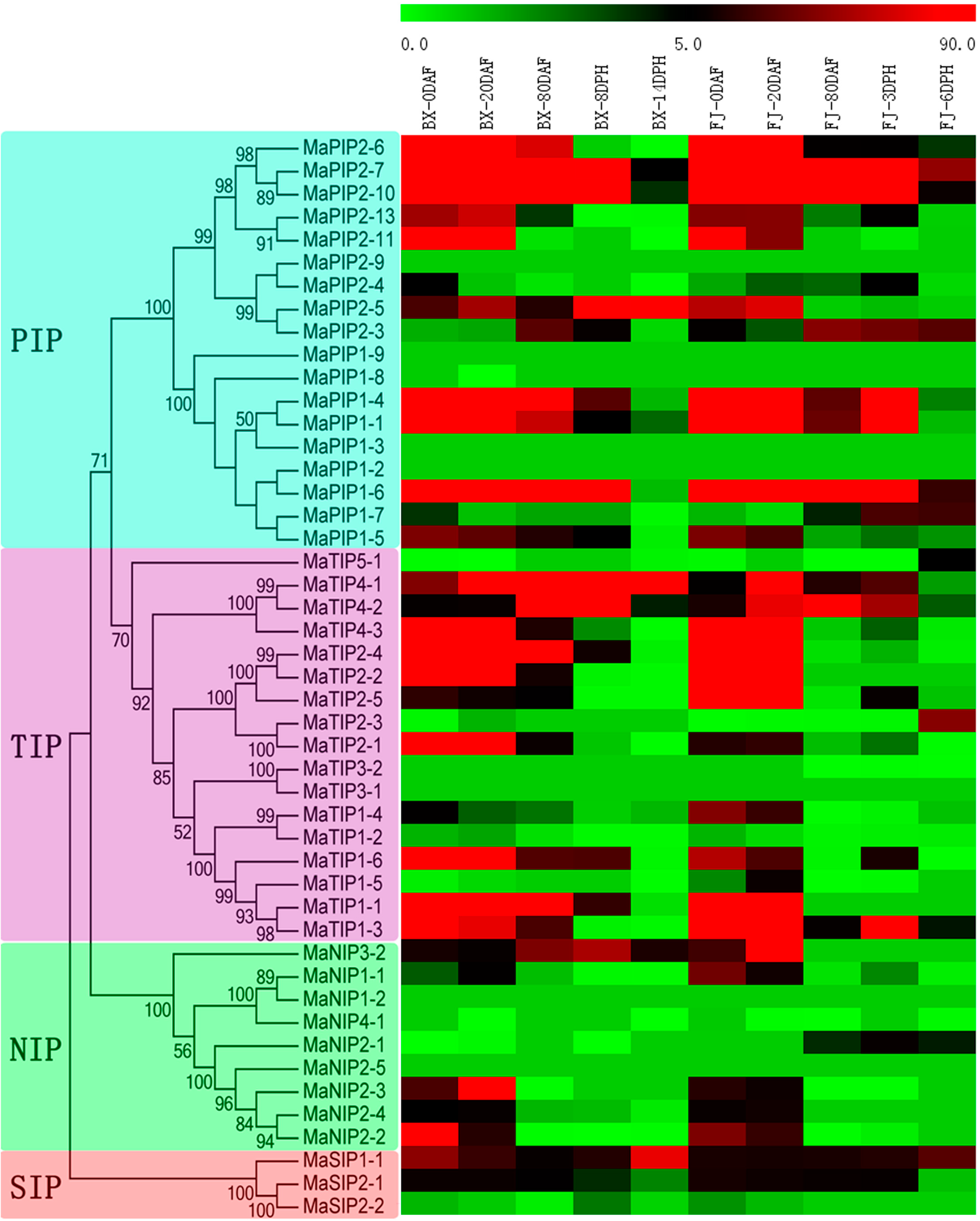
2.7. Expression Analysis of MaAQP Genes in Response to Cold, Salt, and Osmotic Stresses in Two Banana Varieties

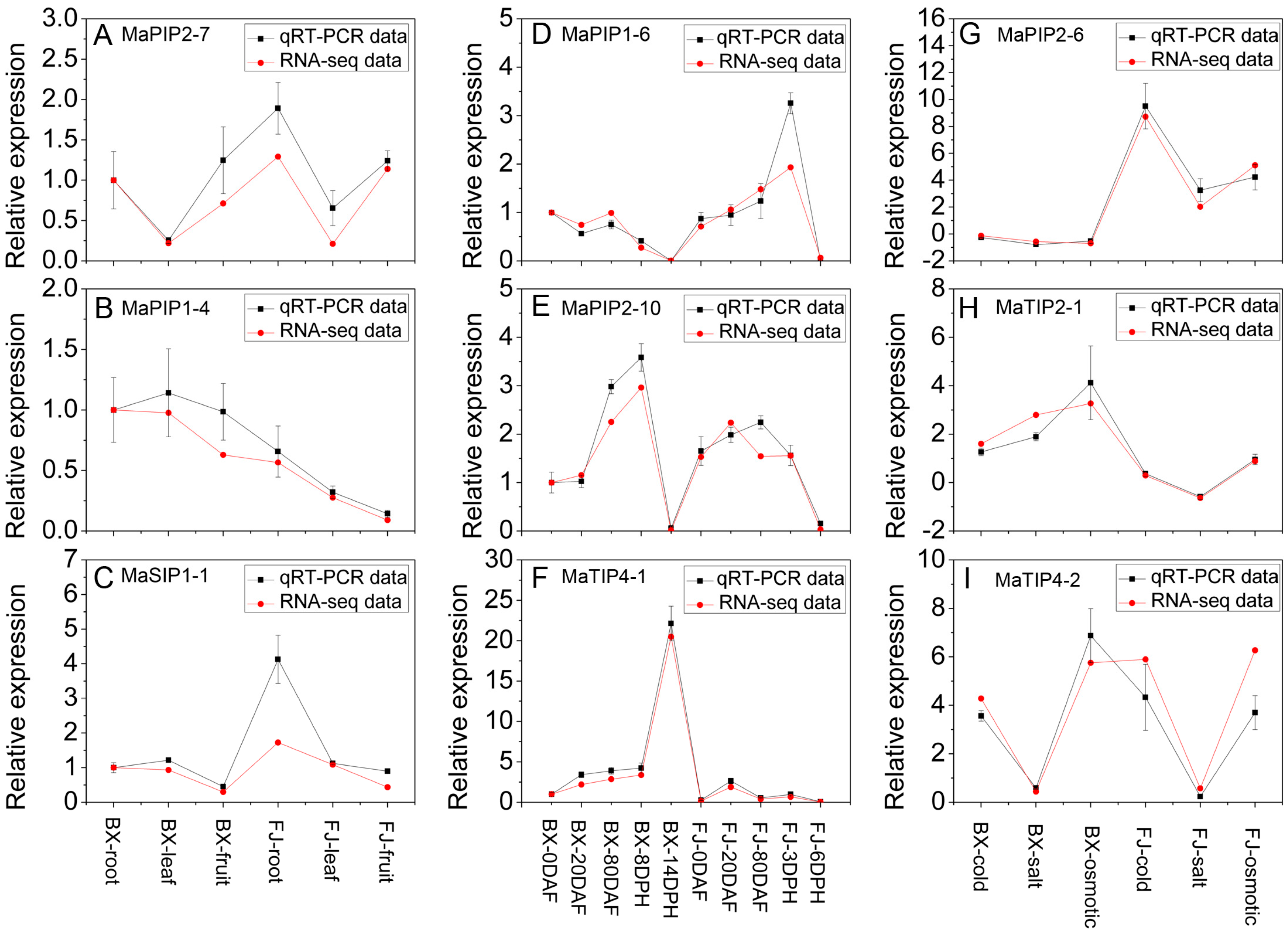
2.8. Validation of the Differentially Expressed AQP Genes by qRT-PCR Analysis
3. Discussion
3.1. Identification and Evolutionary Analysis of MaAQPs
3.2. AQP Genes Are Involved in Fruit Development and Postharvest Ripening of Banana
3.3. AQP Genes Are Involved in Banana Responses to Abiotic Stress
4. Experimental Section
4.1. Plant Materials and Treatments
4.2. Identification and Phylogenetic Analyses
4.3. Protein Properties and Sequence Analyses
4.4. Transcriptome Analysis
4.5. qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Uehlein, N.; Lovisolo, C.; Siefritz, F.; Kaldenhoff, R. The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 2003, 425, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Kaldenhoff, R.; Fischer, M. Aquaporins in plants. Acta Physiol. 2006, 187, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Sade, N.; Gebretsadik, M.; Seligmann, R.; Schwartz, A.; Wallach, R.; Moshelion, M. The role of tobacco aquaporin1 in improving water use efficiency, hydraulic conductivity, and yield production under salt stress. Am. Soc. Plant Physiol. 2010, 152, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Baiges, I.; Schäffner, A.R.; Affenzeller, M.J.; Mas, A. Plant aquaporins. Physiol Plant. 2002, 115, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Tao, P.; Zhong, X.; Li, B.; Wang, W.; Yue, Z.; Lei, J.; Guo, W.; Huang, X. Genome-wide identification and characterization of aquaporin genes (AQPs) in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genom. 2014, 289, 1131–1145. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, Z.Y.; Lin, H.; Cui, W.E.; Chen, J.; Liu, M.; Chen, Z.L.; Qu, L.J.; Gu, H. Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 2006, 16, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Johanson, U.; Karlsson, M.; Johansson, I.; Gustavsson, S.; Jovall, S.; Fraysse, F.; Weig, A.R.; Kjellbom, P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Am. Soc. Plant Physiol. 2001, 126, 1358–1369. [Google Scholar] [CrossRef]
- Kammerloher, W.; Fischer, U.; Piechotta, G.P.; Schäffner, A.R. Water channels in the plant plasma membrane cloned by immunoselection from an expression system. Plant J. 1994, 6, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Johansson, I.; Bush, M.; McCann, M.C.; Maurel, C.; Larsson, C. An abundant TIP expressed in mature highly vacuolated cells. Plant J. 2000, 21, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Weaver, C.D.; Crombie, B.; Stacey, G.; Roberts, D.M. Calcium-dependent phosphorylation of symbiosome membrane proteins from nitrogen-fixing soybean nodules: Evidence for phosphorylation of nodulin-26. Am. Soc. Plant Physiol. 1991, 95, 222–227. [Google Scholar] [CrossRef]
- Chaumont, F.; Barrieu, F.; Wojcik, E.; Chrispeels, M.J.; Jung, R. Aquaporins constitute a large and highly divergent protein family in maize. Am. Soc. Plant Physiol. 2001, 125, 1206–1215. [Google Scholar] [CrossRef]
- Gustavsson, S.; Lebrun, A.S.; Norden, K.; Chaumont, F.; Johanson, U. A novel plant major intrinsic protein in Physcomitrella patens most similar to bacterial glycerol channels. Am. Soc. Plant Physiol. 2005, 139, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Danielson, J.A.H.; Johanson, U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 2008, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Lopez, D.; Bronner, G.; Brunel, N.; Auguin, D.; Bourgerie, S.; Brignolas, F.; Carpin, S.; Tournaire-Roux, C.; Maurel, C.; Fumanal, B.; et al. Insights into Populus XIP aquaporins: Evolutionary expansion, protein functionality, and environmental regulation. J. Exp. Bot. 2012, 63, 2217–2230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreedharan, S.; Shekhawat, U.K.; Ganapathi, T.R. Transgenic banana plants overexpressing a native plasma membrane aquaporin MusaPIP1;2 display high tolerance levels to different abiotic stresses. Plant Biotechnol. J. 2013, 11, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Wallace, I.S.; Roberts, D.M. Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Am. Soc. Plant Physiol. 2004, 135, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.B.; Sankararamakrishnan, R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: Characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 2009, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Mitani-Ueno, N.; Yamaji, N.; Zhao, F.; Ma, J.F. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J. Exp. Bot. 2011, 62, 4391–4398. [Google Scholar] [CrossRef] [PubMed]
- Eisenbarth, D.A.; Weig, A.R. Dynamics of aquaporins and water relations during hypocotyl elongation in Ricinus communis L. seedlings. J. Exp. Bot. 2005, 56, 1831–1842. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Sawa, Y.; Ishikawa, T.; Shibata, H. Phosphorylation of plasma membrane aquaporin regulates temperature-dependent opening of tulip petals. Plant Cell Physiol. 2004, 45, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Ruan, X.M.; Zhang, J.; Wu, Y.J.; Wang, X.L.; Li, X.B. Cotton plasma membrane intrinsic protein 2s (PIP2s) selectively interact to regulate their water channel activities and are required for fibre development. New Phytol. 2013, 199, 695–707. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Hu, W.; Deng, X.; Ma, Z.; Chen, L.; Huang, C.; Wang, C.; Wang, J.; He, Y.; Yang, G.; et al. Overexpression of the wheat aquaporin gene, TaAQP7, enhances drought tolerance in transgenic tobacco. PLoS ONE 2012, 7, e52439. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yuan, Q.; Wang, Y.; Cai, R.; Deng, X.; Wang, J.; Zhou, S.; Chen, M.; Chen, L.; Huang, C.; et al. Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol. 2012, 53, 2127–2141. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, W.; Liu, J.; Zhang, J.; Jia, C.; Miao, H.; Xu, B.; Jin, Z. A banana aquaporin gene, MaPIP1;1, is involved in tolerance to drought and salt stresses. BMC Plant Biol. 2014, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Sreedharan, S.; Shekhawat, U.K.; Ganapathi, T.R. Constitutive and stress-inducible overexpression of a native aquaporin gene (MusaPIP2;6) in transgenic banana plants signals its pivotal role in salt tolerance. Plant Mol. Biol. 2015, 88, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, J.; Ishikawa, F.; Yamaguchi, T.; Uemura, M.; Maeshima, M. Identifiation of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005, 46, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.X.; Moon, S.; Jung, K.H. Genome–wide expression analysis of rice aquaporin genes and development of a functional gene network mediated by aquaporin expression in roots. Planta 2013, 238, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Reuscher, S.; Akiyama, M.; Mori, C.; Aoki, K.; Shibata, D.; Shiratake, K. Genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE 2013, 8, e79052. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Lovato, A.; Polverari, A.; Wang, M.; Liang, Y.H.; Ma, Y.C.; Cheng, Z.M. Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera). BMC Plant Biol. 2014, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Am. Soc. Plant Physiol. 2003, 132, 530–543. [Google Scholar]
- Ariani, A.; Gepts, P. Genome-wide identification and characterization of aquaporin gene family in common bean (Phaseolus vulgaris L.). Mol. Genet. Genom. 2015. [Google Scholar] [CrossRef]
- Venkatesh, J.; Yu, J.W.; Park, S.W. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol. Biochem. 2013, 73, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Santoni, V.; Maurel, C. Plant aquaporins: Role in plant physiology. Biochim. Biophys. Acta 2014, 1840, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Heckwolf, M.; Pater, D.; Hanson, D.T.; Kaldenhoff, R. The Arabidopsis thaliana aquaporin AtPIP1;2 is a physiologically relevant CO2 transport facilitator. Plant J. 2011, 67, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Mori, I.C.; Rhee, J.; Shibasaka, M.; Sasano, S.; Kaneko, T.; Horiem, T.; Katsuharam, M. CO2 transport by PIP2 aquaporins in barley. Plant Cell Physiol. 2014, 55, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ludewig, U.; Gasset, B.; Frommer, W.B.; von Wiren, N. Urea transport by nitrogen-regulated tonoplastic intrinsic proteins in Arabidopsis. Am. Soc. Plant Physiol. 2003, 133, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.M.; Jahn, T.P.; Møller, A.L.; Schjoerring, J.K.; Ferri, D.; Klaerke, D.A.; Zeuthen, T. NH3 and NH4+ permeability in aquaporin expressin Xenopus oocytes. Pflg. Arch. 2005, 450, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Loque, D.; Ludewig, U.; Yuan, L.; von Wiren, N. Tonoplastic intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Am. Soc. Plant Physiol. 2005, 137, 671–680. [Google Scholar] [CrossRef]
- Bienert, G.P.; Møller, A.L.; Kristiansen, K.A.; Schulz, A.; Møller, I.M.; Schjoerring, J.K.; Jahn, T.P. Specifi aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol. Chem. 2007, 282, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.M.; Rivers, R.L.; Zeidel, M.L.; Roberts, D.M. Purifiation and functional reconstitution of soybean nodulin 26. An aquaporin with water and glycerol transport properties. Biochemistry 1999, 38, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Wallace, I.S.; Roberts, D.M. Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry 2005, 44, 16826–16834. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.G.; Tamai, K.; Yamaji, N.; Mitani, N.; Konishi, S.; Katsuhara, M.; Ishiguro, M.; Murata, Y.; Yano, M. A silicon transporter in rice. Nature 2006, 440, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Takano, J.; Wada, M.; Ludewig, U.; Schaaf, G.; von Wirén, N.; Fujiwara, T. The Arabidopsis major intrinsic protein NIP5;1 is essential for effiient boron uptake and plant development under boron limitation. Plant Cell. 2006, 18, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.G.; Roberts, D.M. Arabidopsis NIP2;1: A major intrinsic protein transporter of lactic acid induced by anoxic stress. J. Biol. Chem. 2007, 282, 24209–24218. [Google Scholar] [CrossRef] [PubMed]
- Bapat, V.A.; Trivedi, P.K.; Ghosh, A.; Sane, V.A.; Ganapathi, T.R.; Nath, P. Ripening of fleshy fruit: Molecular insight and the role of ethylene. Biotechnol. Adv. 2010, 28, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Choat, B.; Gambetta, G.A.; Shackel, K.A.; Matthews, M.A. Vascular function in grape berries across development and its relevance to apparent hydraulic isolation. Am. Soc. Plant Physiol. 2009, 151, 1677–1687. [Google Scholar] [CrossRef] [PubMed]
- Alleva, K.; Marquez, M.; Villarreal, N.; Mut, P.; Bustamante, C.; Bellati, J.; Martínez, G.; Civello, M.; Amodeo, G. Cloning, functional characterization, and co-expression studies of a novel aquaporin (FaPIP2;1) of strawberry fruit. J. Exp. Bot. 2010, 61, 3935–3945. [Google Scholar] [CrossRef] [PubMed]
- Vera-Estrella, R.; Barkla, B.J.; Bohnert, H.J.; Pantoja, O. Novel regulation of aquaporins during osmotic stress. Am. Soc. Plant Physiol. 2004, 135, 2318–2329. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Liu, J.H.; Yang, X.Y.; Zhang, J.B.; Jia, C.H.; Li, M.Y.; Xu, B.Y.; Jin, Z.Q. Identification of a gene encoding glutamate decarboxylase involved in postharvest fruit ripening process in banana. HortScience 2014, 49, 1056–1060. [Google Scholar]
- Sun, Q.; Zhang, N.; Wang, J.; Zhang, H.; Li, D.; Shi, J.; Li, R.; Weeda, S.; Zhao, B.; Ren, S.; et al. Melatonin promotes ripening and improves quality of tomato fruit during postharvest life. J. Exp. Bot. 2015, 66, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Van Asten, P.J.A.; Fermont, A.M.; Taulya, G. Drought is a major yield loss factor for rainfed East African highland banana. Agric. Water Manag. 2011, 98, 541–552. [Google Scholar] [CrossRef]
- Gao, Z.X.; He, X.L.; Zhao, B.C.; Zhou, C.J.; Liang, Y.Z.; Ge, R.C.; Shen, Y.; Huang, Z. Overexpressing a putative aquaporin gene from wheat, TaNIP, enhances salt tolerance in transgenic Arabidopsis. Plant Cell Physiol. 2010, 51, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.W.; Gudimella, R.; Harikrishna, J.A.; Sin, L.W.; Khalid, N.; Keulemans, J. A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genom. 2013, 14, 683. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.C.; Xiao, Y.; Zhao, C.X. Some physiological responses of banana to water stress. J. Zhongkai Agrotech. Coll. 1999, 12, 40–42. [Google Scholar]
- Bananuka, J.A.; Rubaihayo, P.R.; Tenywa, M.M. Reactions of Musa genotypes to drought stress. Afr. Crop Sci. J. 1999, 7, 333–339. [Google Scholar]
- Pua, E.C.; Chandramouli, S.; Han, P.; Liu, P. Malate synthase gene expression during fruit ripening of Cavendish banana (Musa acuminata cv. Williams). J. Exp. Bot. 2003, 54, 309–316. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Baxter, L.; Hickman, R.; Beynon, J.; Moore, J.D.; Ott, S. MEME-LaB: Motif analysis in clusters. Bioinformatics 2013, 29, 1696–1697. [Google Scholar] [CrossRef] [PubMed]
- Mulder, N.; Apweiler, R. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol. Biol. 2007, 396, 59–70. [Google Scholar] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Pollier, J.; Rombauts, S.; Goossens, A. Analysis of RNA-Seq data with TopHat and Cufflinks for genome-wide expression analysis of jasmonate-treated plants and plant cultures. Methods Mol. Biol. 2013, 1011, 305–315. [Google Scholar] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, H.Y.; Kuang, J.F.; Li, J.G.; Lu, W.J.; Chen, J.Y. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time Quantitative PCR and the 2–ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; Hou, X.; Huang, C.; Yan, Y.; Tie, W.; Ding, Z.; Wei, Y.; Liu, J.; Miao, H.; Lu, Z.; et al. Genome-Wide Identification and Expression Analyses of Aquaporin Gene Family during Development and Abiotic Stress in Banana. Int. J. Mol. Sci. 2015, 16, 19728-19751. https://doi.org/10.3390/ijms160819728
Hu W, Hou X, Huang C, Yan Y, Tie W, Ding Z, Wei Y, Liu J, Miao H, Lu Z, et al. Genome-Wide Identification and Expression Analyses of Aquaporin Gene Family during Development and Abiotic Stress in Banana. International Journal of Molecular Sciences. 2015; 16(8):19728-19751. https://doi.org/10.3390/ijms160819728
Chicago/Turabian StyleHu, Wei, Xiaowan Hou, Chao Huang, Yan Yan, Weiwei Tie, Zehong Ding, Yunxie Wei, Juhua Liu, Hongxia Miao, Zhiwei Lu, and et al. 2015. "Genome-Wide Identification and Expression Analyses of Aquaporin Gene Family during Development and Abiotic Stress in Banana" International Journal of Molecular Sciences 16, no. 8: 19728-19751. https://doi.org/10.3390/ijms160819728





