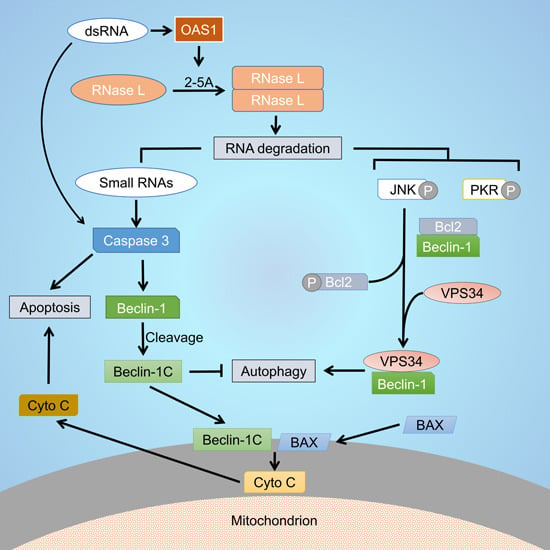
RNase L Cleavage Products Promote Switch from Autophagy to Apoptosis by Caspase-Mediated Cleavage of Beclin-1
Abstract
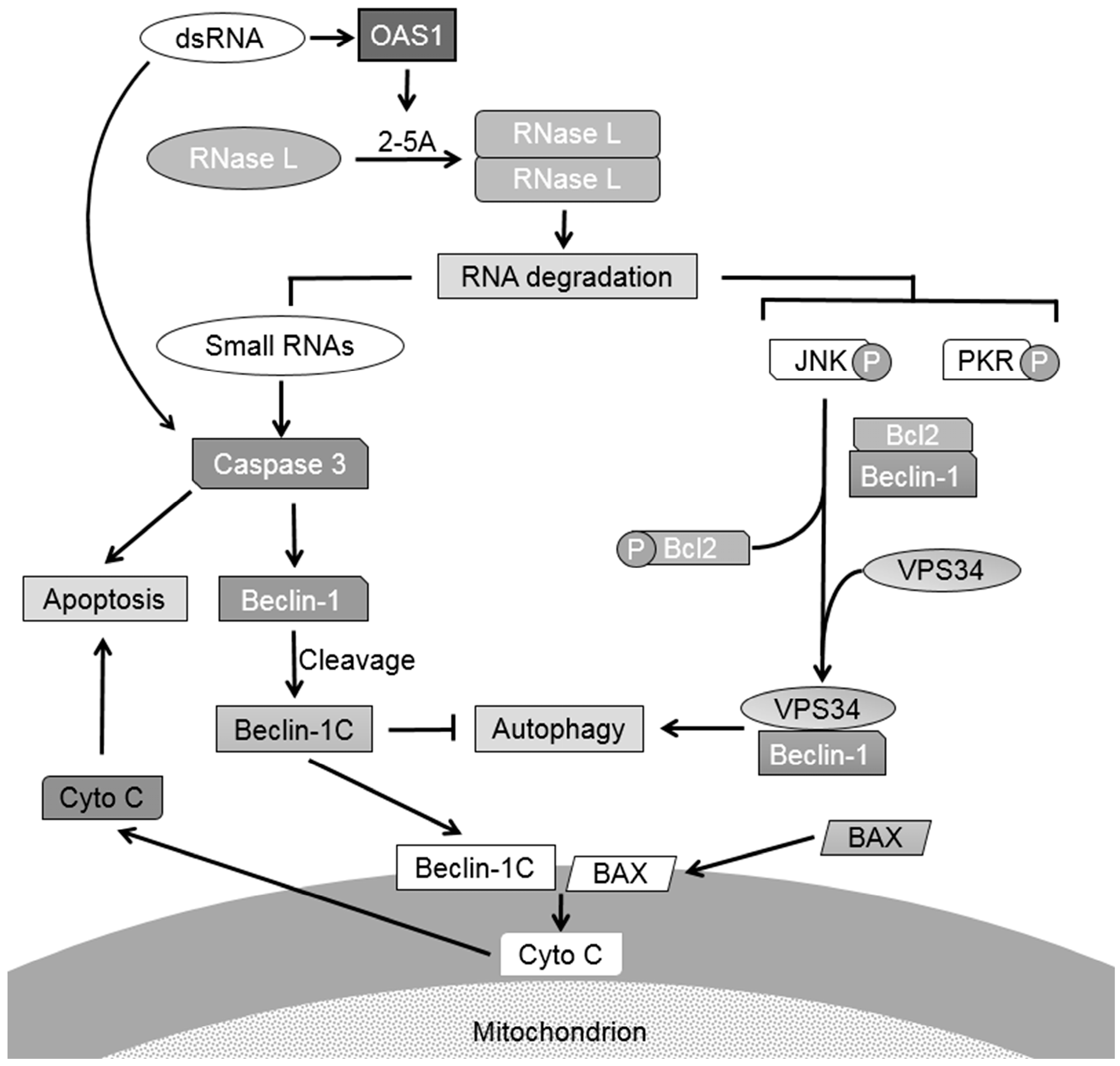
:1. Introduction
2. Results
2.1. RNase L and dsRNA Signaling Pathways Regulate Cross-Talk between Autophagy and Apoptosis
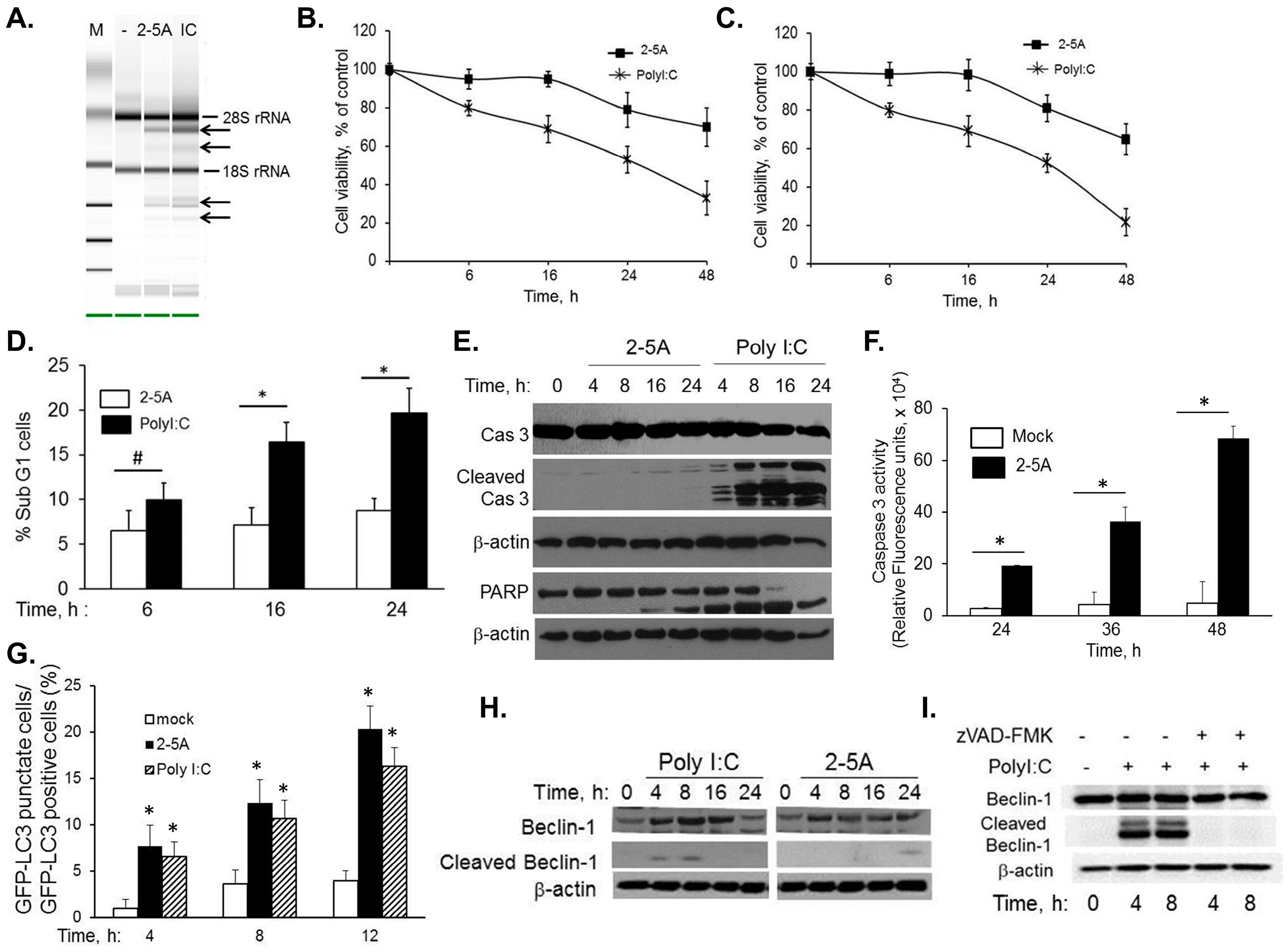
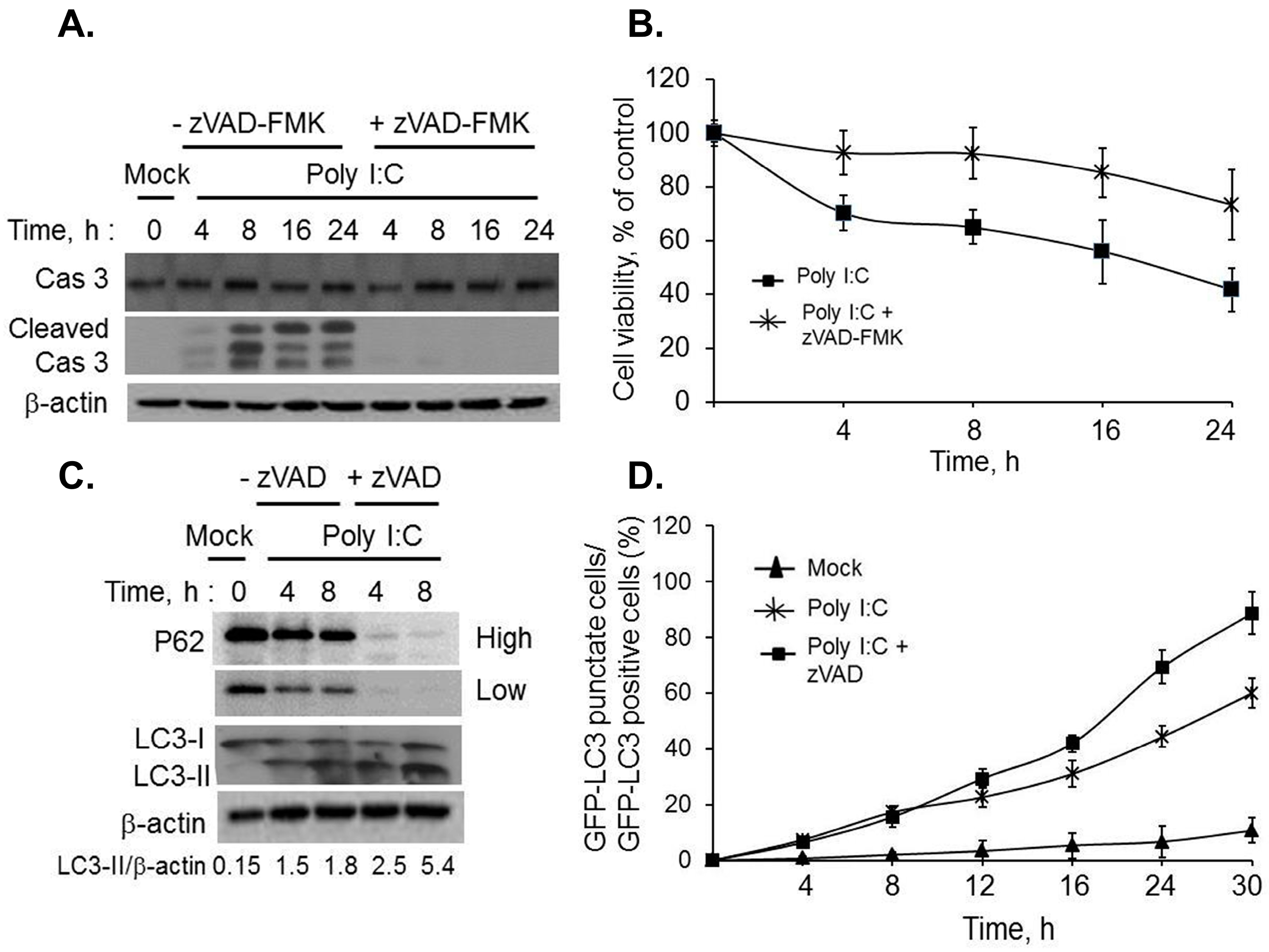
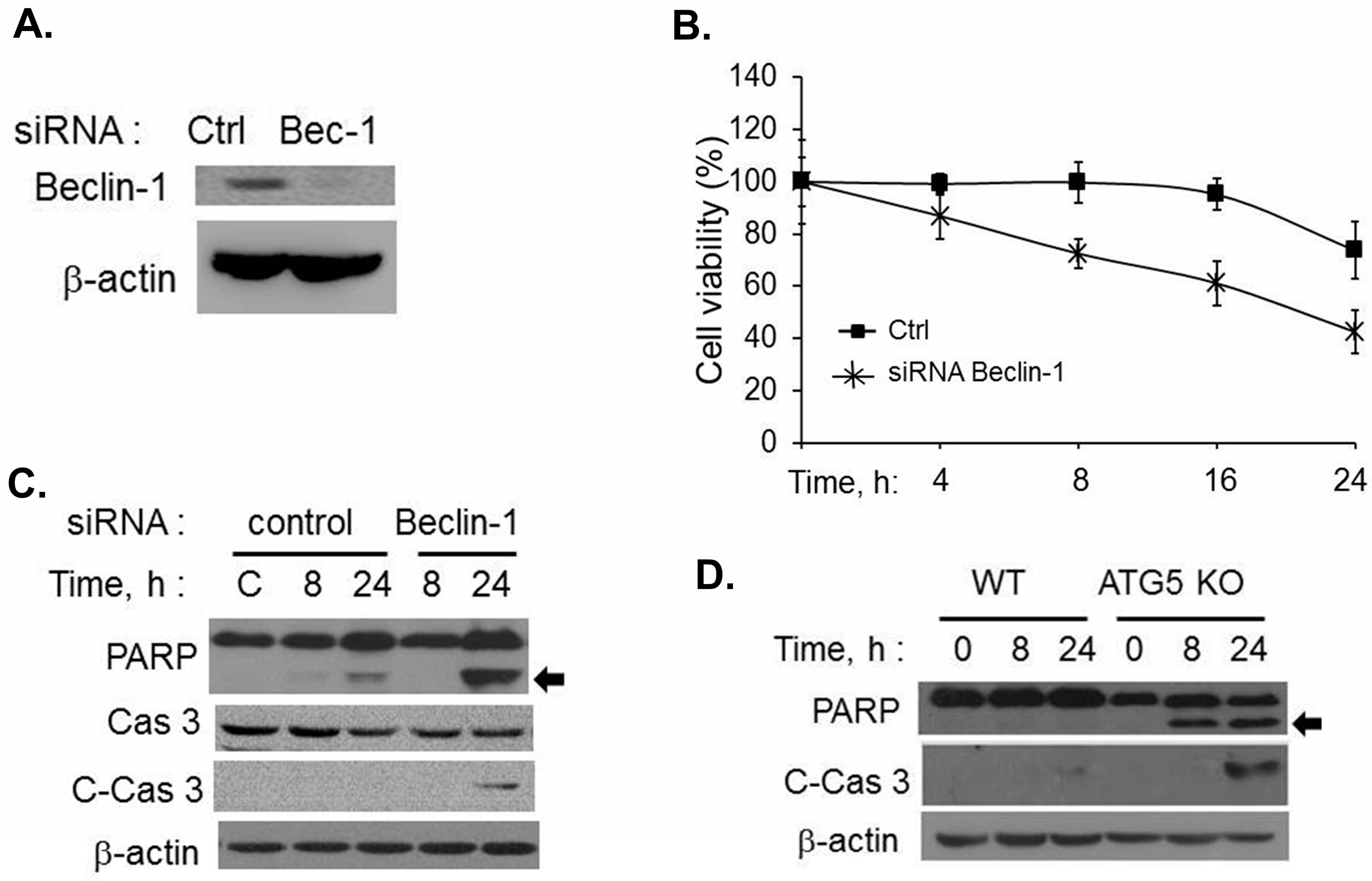
2.2. Inhibiting RNase L-Induced Autophagy Leads to Apoptosis by Cleavage of Beclin-1
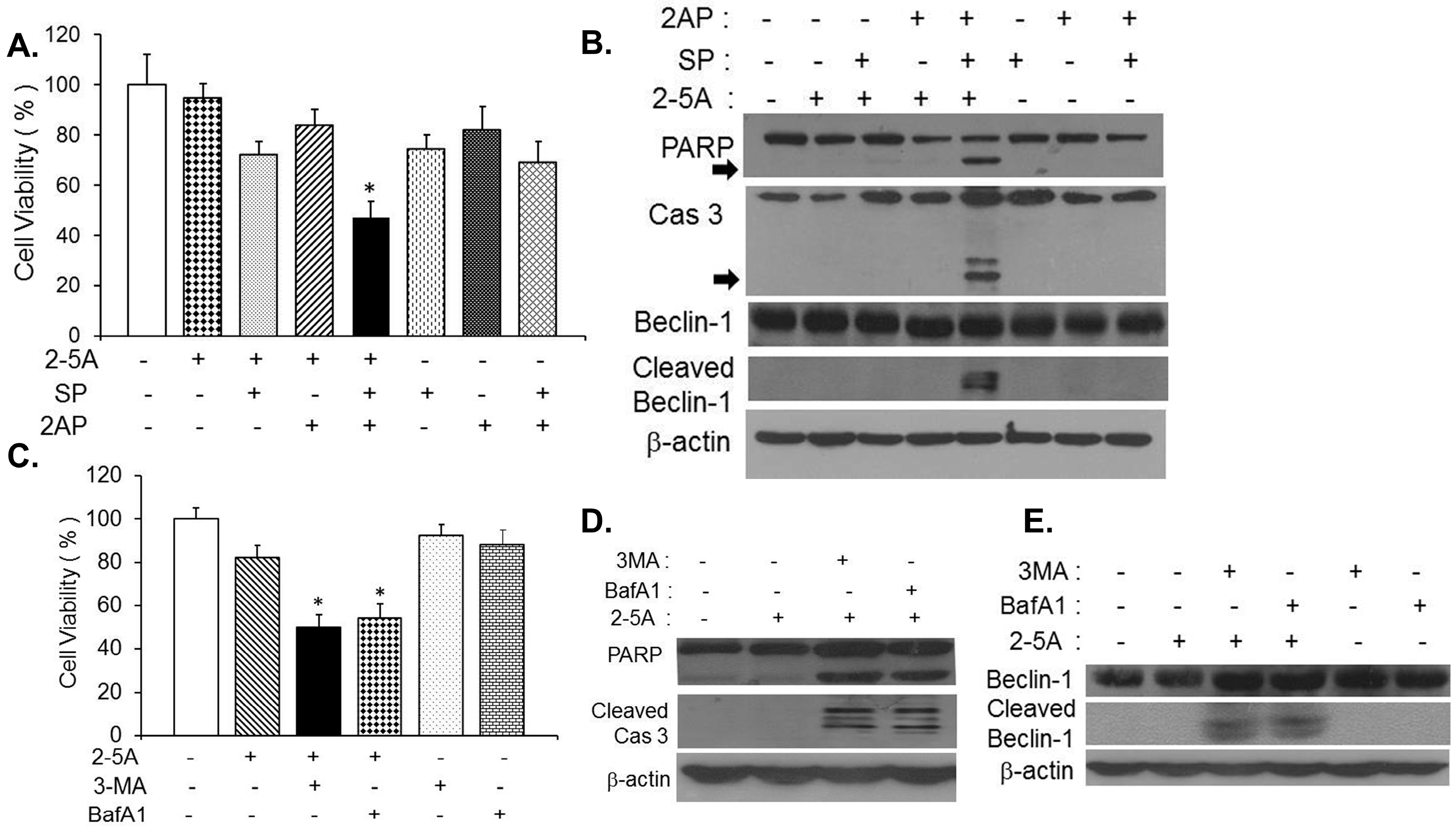
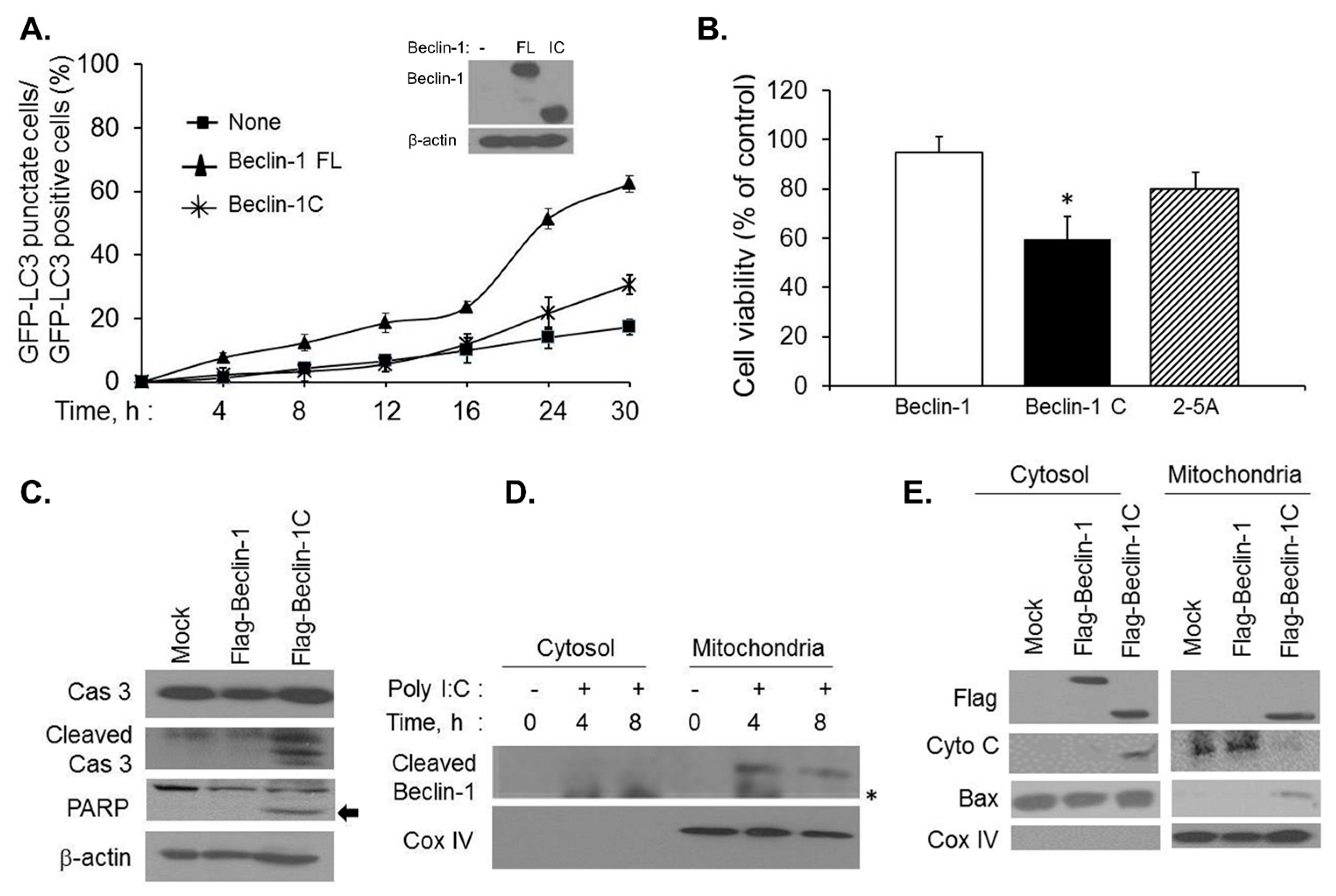
2.3. Caspase-Cleaved Beclin-1 Fragments Induce Apoptosis by Translocation of Bax to the Mitochondria and Release of Cytochrome c
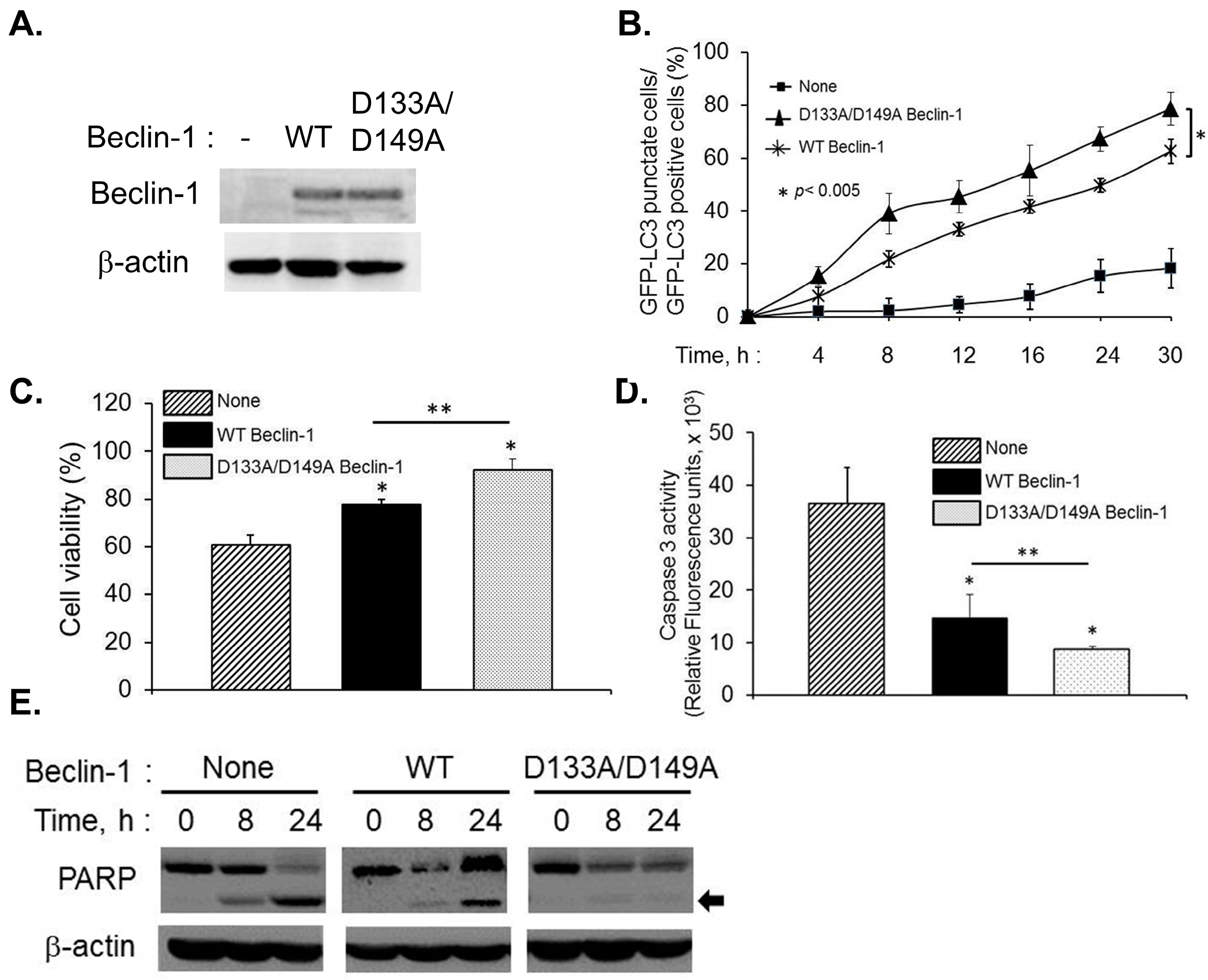
2.4. Caspase-Cleavage Resistant Beclin-1 Inhibits Apoptosis Induction and Prolongs Autophagy
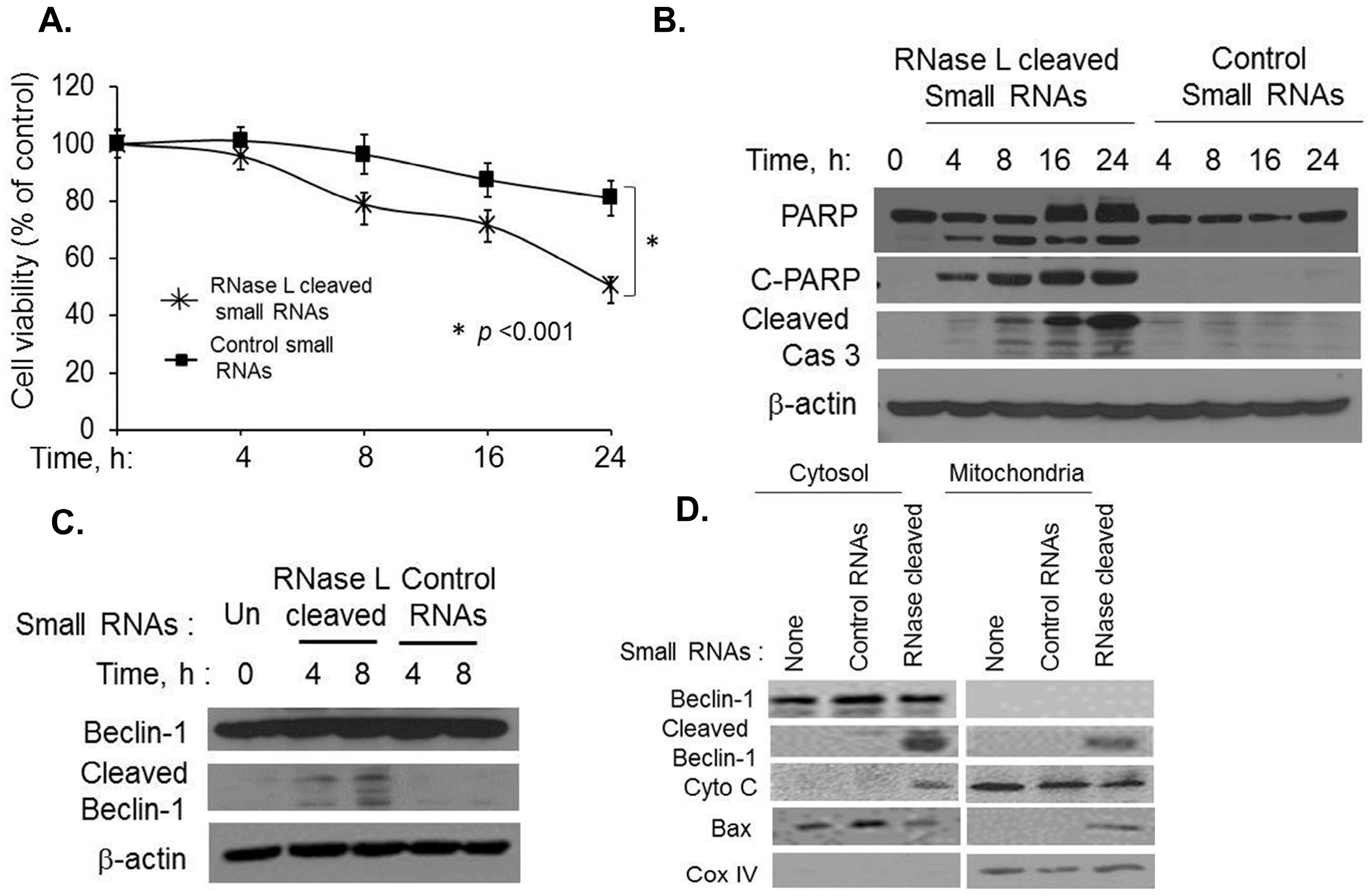
2.5. Small dsRNA Cleavage Products of RNase L Activity Modulate Switch from Autophagy to Apoptosis
3. Discussion
4. Experimental Section
4.1. Chemicals, Reagents and Antibodies
4.2. Plasmids
| pcDNA4-Beclin1-C | 5ʹ-CGCGGATCCATGACTCAGCTCAACGTCACTGAAAATGAG-3ʹ (Forward) 5ʹ-CCGCTCGAGTCACTTGTCATCGTCATCC-3ʹ (Reverse) |
| pcDNA4-Beclin-1-D133A-D149A | D133A-forward: 5ʹCAGTGACGTTGAGCTGAGTAGCCAGCTGGTCTAAAAGAGT-3ʹ D133A-reverse: 5ʹTCACAGAGTGGGTGAGCCACATCTGTCTGGC-3ʹ D149A-forward: 5ʹACTCTTTTAGACCAGCTGGCTACTCAGCTCAACGTCACTG-3ʹ D149A-reverse: 5ʹGCCAGACAGATGTGGCTCACCCACTCTGTGA-3ʹ |
4.3. Cell Culture and Transfections
4.4. Monitoring RNase L Activity in Cells and Isolation and Purification of RNase L Cleaved RNAs
4.5. Quantification of Autophagy and Immunofluorescence
4.6. Immunoblot Analysis
4.7. Cell Fractionation
4.8. Cell Viability Assays
4.9. Caspase 3/7 Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Gordy, C.; He, Y.W. The crosstalk between autophagy and apoptosis: Where does this lead? Protein Cell 2012, 3, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Levine, B. The autophagy effector Beclin 1: A novel BH3-only protein. Oncogene 2008, 27, S137–S148. [Google Scholar] [CrossRef] [PubMed]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Levine, B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010, 22, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Le Toumelin, G.; Criollo, A.; Rain, J.C.; Gautier, F.; Juin, P.; Tasdemir, E.; Pierron, G.; Troulinaki, K.; Tavernarakis, N.; et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007, 26, 2527–2539. [Google Scholar] [CrossRef] [PubMed]
- Oberstein, A.; Jeffrey, P.D.; Shi, Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 2007, 282, 13123–13132. [Google Scholar] [CrossRef] [PubMed]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, A.D.; Eisenstein, M.; Ber, Y.; Bialik, S.; Kimchi, A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell 2011, 44, 698–709. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.A.; Malathi, K. RNase L induces autophagy via c-Jun N-terminal kinase and double-stranded RNA-dependent protein kinase signaling pathways. J. Biol. Chem. 2012, 287, 43651–43664. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Pattingre, S.; Sinha, S.; Bassik, M.; Levine, B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 2008, 30, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Sinha, S.; Levine, B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 2008, 4, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Zalckvar, E.; Berissi, H.; Mizrachy, L.; Idelchuk, Y.; Koren, I.; Eisenstein, M.; Sabanay, H.; Pinkas-Kramarski, R.; Kimchi, A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009, 10, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, E.; Vande Walle, L.; Kersse, K.; Cornelis, S.; Claerhout, S.; Vanoverberghe, I.; Roelandt, R.; de Rycke, R.; Verspurten, J.; Declercq, W.; et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010, 1, e18. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Rubinsztein, D.C. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: An effect rescued by Bcl-xL. Cell Death Differ. 2010, 17, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.H.; Jo, Y.K.; Hwang, J.J.; Lee, Y.M.; Roh, S.A.; Kim, J.C. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 2009, 274, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Djavaheri-Mergny, M.; Maiuri, M.C.; Kroemer, G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010, 29, 1717–1719. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, P.; Sun, Q.; Ding, W.X.; Yin, X.M.; Sobol, R.W.; Stolz, D.B.; Yu, J.; Zhang, L. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res. 2011, 71, 3625–3634. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Han, J.; Lu, C.; Goldstein, L.A.; Rabinowich, H. Autophagic degradation of active caspase-8: A crosstalk mechanism between autophagy and apoptosis. Autophagy 2010, 6, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Li, Q.; Lee, J.Y.; Lee, S.H.; Jeong, J.H.; Lee, H.R.; Chang, H.; Zhou, F.C.; Gao, S.J.; Liang, C.; et al. FLIP-mediated autophagy regulation in cell death control. Nat. Cell Biol. 2009, 11, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Jounai, N.; Takeshita, F.; Kobiyama, K.; Sawano, A.; Miyawaki, A.; Xin, K.Q.; Ishii, K.J.; Kawai, T.; Akira, S.; Suzuki, K.; et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14050–14055. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, S.; Perozzo, R.; Schmid, I.; Ziemiecki, A.; Schaffner, T.; Scapozza, L.; Brunner, T.; Simon, H.U. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006, 8, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Hou, W.; Goldstein, L.A.; Stolz, D.B.; Watkins, S.C.; Rabinowich, H. A complex between Atg7 and Caspase-9: A novel mechanism of cross-regulation between autophagy and apoptosis. J. Biol. Chem. 2014, 289, 6485–6497. [Google Scholar] [CrossRef] [PubMed]
- Borden, E.C.; Sen, G.C.; Uze, G.; Silverman, R.H.; Ransohoff, R.M.; Foster, G.R.; Stark, G.R. Interferons at age 50: Past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007, 6, 975–990. [Google Scholar] [CrossRef] [PubMed]
- Akira, S. Pathogen recognition by innate immunity and its signaling. Proc. Jpn. Acad. Seri. B Phys. Biol. Sci. 2009, 85, 143–156. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Gantier, M.P.; Williams, B.R. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007, 18, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Gantier, M.P.; Williams, B.R. Making sense of viral RNA sensing. Mol. Ther.: J. Am. Soc. Gene Ther. 2011, 19, 1578–1581. [Google Scholar] [CrossRef] [PubMed]
- Barber, G.N. The dsRNA-dependent protein kinase, PKR and cell death. Cell Death Differ. 2005, 12, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Der, S.D.; Yang, Y.L.; Weissmann, C.; Williams, B.R. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 1997, 94, 3279–3283. [Google Scholar] [CrossRef] [PubMed]
- McAllister, C.S.; Samuel, C.E. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J. Biol. Chem. 2009, 284, 1644–1651. [Google Scholar] [CrossRef] [PubMed]
- Besch, R.; Poeck, H.; Hohenauer, T.; Senft, D.; Hacker, G.; Berking, C.; Hornung, V.; Endres, S.; Ruzicka, T.; Rothenfusser, S.; et al. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J. Clin. Investig. 2009, 119, 2399–2411. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, O.; Ali, M.M.; Luo, S.S.; Ohba, T.; Katabuchi, H.; Takeshita, T.; Takizawa, T. Short RNA duplexes elicit RIG-I-mediated apoptosis in a cell type- and length-dependent manner. Sci. Signal. 2011, 4, ra74. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Esteban, M. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): Mechanism of action. Apoptosis: An Int. J. Program. Cell Death 2000, 5, 107–114. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Marques, J.T.; Yamashita, M.; Peters, K.L.; Smith, K.; Desai, A.; Williams, B.R.; Sen, G.C. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 2010, 29, 1762–1773. [Google Scholar] [CrossRef] [PubMed]
- Vitali, P.; Scadden, A.D. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct. Mol. Biol. 2010, 17, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Kerr, I.M.; Wreschner, D.H.; Silverman, R.H.; Cayley, P.J.; Knight, M. The 2–5A (pppA2ʹp5ʹA2ʹp5ʹA) and protein kinase systems in interferon-treated and control cells. Adv. Cyclic Nucleotide Res. 1981, 14, 469–478. [Google Scholar] [PubMed]
- Zhou, A.; Hassel, B.A.; Silverman, R.H. Expression cloning of 2-5A-dependent RNAase: A uniquely regulated mediator of interferon action. Cell 1993, 72, 753–765. [Google Scholar] [CrossRef]
- Silverman, R.H.; Cayley, P.J.; Knight, M.; Gilbert, C.S.; Kerr, I.M. Control of the ppp(a2ʹp)nA system in HeLa cells. Effects of interferon and virus infection. Eur. J. Biochem./FEBS 1982, 124, 131–138. [Google Scholar] [CrossRef]
- Wreschner, D.H.; James, T.C.; Silverman, R.H.; Kerr, I.M. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2'p)nA) in interferon-treated cells. Nucleic Acids Res. 1981, 9, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Malathi, K.; Dong, B.; Gale, M.; Silverman, R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Banerjee, S.; Franchi, L.; Loo, Y.M.; Gale, M; Nunez, G.; Silverman, R.H. RNase L Activates the NLRP3 Inflammasome during Viral Infections. Cell Host Microbe 2015, 17, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Xiang, Y.; Sabapathy, K.; Silverman, R.H. An apoptotic signaling pathway in the interferon antiviral response mediated by RNase L and c-Jun NH2-terminal kinase. J. Biol. Chem. 2004, 279, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Malathi, K.; Paranjape, J.M.; Ganapathi, R.; Silverman, R.H. HPC1/RNASEL mediates apoptosis of prostate cancer cells treated with 2ʹ,5ʹ-oligoadenylates, topoisomerase I inhibitors, and tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2004, 64, 9144–9151. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Chakrabarti, A.; Jha, B.K.; Weiss, S.R.; Silverman, R.H. Cell-type-specific effects of RNase L on viral induction of beta interferon. mBio 2014, 5, e00856–e00814. [Google Scholar] [CrossRef] [PubMed]
- Silverman, R.H.; Skehel, J.J.; James, T.C.; Wreschner, D.H.; Kerr, I.M. rRNA cleavage as an index of ppp(A2ʹp)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J. Virol. 1983, 46, 1051–1055. [Google Scholar] [PubMed]
- Malathi, K.; Paranjape, J.M.; Bulanova, E.; Shim, M.; Guenther-Johnson, J.M.; Faber, P.W.; Eling, T.E.; Williams, B.R.; Silverman, R.H. A transcriptional signaling pathway in the IFN system mediated by 2ʹ-5ʹ-oligoadenylate activation of RNase L. Proc. Natl. Acad. Sci. USA 2005, 102, 14533–14538. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhao, L.; Liu, L.; Gao, P.; Tian, W.; Wang, X.; Jin, H.; Xu, H.; Chen, Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 2010, 1, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, S.; Roberts, P.C.; Brown, L.E.; Truong, H.; Pattnaik, A.K.; Archer, D.R.; Barber, G.N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 2000, 13, 129–141. [Google Scholar] [CrossRef]
- Silverman, R.H. Viral encounters with 2ʹ,5ʹ-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007, 81, 12720–12729. [Google Scholar] [CrossRef] [PubMed]
- Rusch, L.; Zhou, A.; Silverman, R.H. Caspase-dependent apoptosis by 2ʹ,5ʹ-oligoadenylate activation of RNase L is enhanced by IFN-beta. J. Interferon Cytokine Res.: J. Int. Soc. Interferon Cytokine Res. 2000, 20, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Booth, L.A.; Tavallai, S.; Hamed, H.A.; Cruickshanks, N.; Dent, P. The role of cell signalling in the crosstalk between autophagy and apoptosis. Cell. Signal. 2014, 26, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Ghosh, P.K.; Banerjee, S.; Gaughan, C.; Silverman, R.H. RNase L triggers autophagy in response to viral infections. J. Virol. 2012, 86, 11311–11321. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, D.Y.; Xu, L.H.; He, X.H.; Zhang, Y.T.; Zeng, L.H.; Cai, J.Y.; Ren, S. Autophagy is differentially induced in prostate cancer LNCaP, DU145 and PC-3 cells via distinct splicing profiles of ATG5. Autophagy 2013, 9, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Talloczy, Z.; Jiang, W.; Virgin, H.W.T.; Leib, D.A.; Scheuner, D.; Kaufman, R.J.; Eskelinen, E.L.; Levine, B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Orvedahl, A.; Alexander, D.; Talloczy, Z.; Sun, Q.; Wei, Y.; Zhang, W.; Burns, D.; Leib, D.A.; Levine, B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 2007, 1, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Colbert, C.L.; Becker, N.; Wei, Y.; Levine, B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy 2008, 4, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.P.; Kirkegaard, K. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 2007, 81, 12543–12553. [Google Scholar] [CrossRef] [PubMed]
- Gannage, M.; Dormann, D.; Albrecht, R.; Dengjel, J.; Torossi, T.; Ramer, P.C.; Lee, M.; Strowig, T.; Arrey, F.; Conenello, G.; et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 2009, 6, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Joubert, P.E.; Werneke, S.W.; de la Calle, C.; Guivel-Benhassine, F.; Giodini, A.; Peduto, L.; Levine, B.; Schwartz, O.; Lenschow, D.J.; Albert, M.L. Chikungunya virus-induced autophagy delays caspase-dependent cell death. J. Exp. Med. 2012, 209, 1029–1047. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Yamaji, T.; Ueno, T.; Ishiura, S.; Kominami, E.; Hanada, K. Consideration about negative controls for LC3 and expression vectors for four colored fluorescent protein-LC3 negative controls. Autophagy 2008, 4, 131–134. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddiqui, M.A.; Mukherjee, S.; Manivannan, P.; Malathi, K. RNase L Cleavage Products Promote Switch from Autophagy to Apoptosis by Caspase-Mediated Cleavage of Beclin-1. Int. J. Mol. Sci. 2015, 16, 17611-17636. https://doi.org/10.3390/ijms160817611
Siddiqui MA, Mukherjee S, Manivannan P, Malathi K. RNase L Cleavage Products Promote Switch from Autophagy to Apoptosis by Caspase-Mediated Cleavage of Beclin-1. International Journal of Molecular Sciences. 2015; 16(8):17611-17636. https://doi.org/10.3390/ijms160817611
Chicago/Turabian StyleSiddiqui, Mohammad Adnan, Sushovita Mukherjee, Praveen Manivannan, and Krishnamurthy Malathi. 2015. "RNase L Cleavage Products Promote Switch from Autophagy to Apoptosis by Caspase-Mediated Cleavage of Beclin-1" International Journal of Molecular Sciences 16, no. 8: 17611-17636. https://doi.org/10.3390/ijms160817611