Development of Ophiocordyceps sinensis through Plant-Mediated Interkingdom Host Colonization
Abstract
:1. Introduction
2. Results
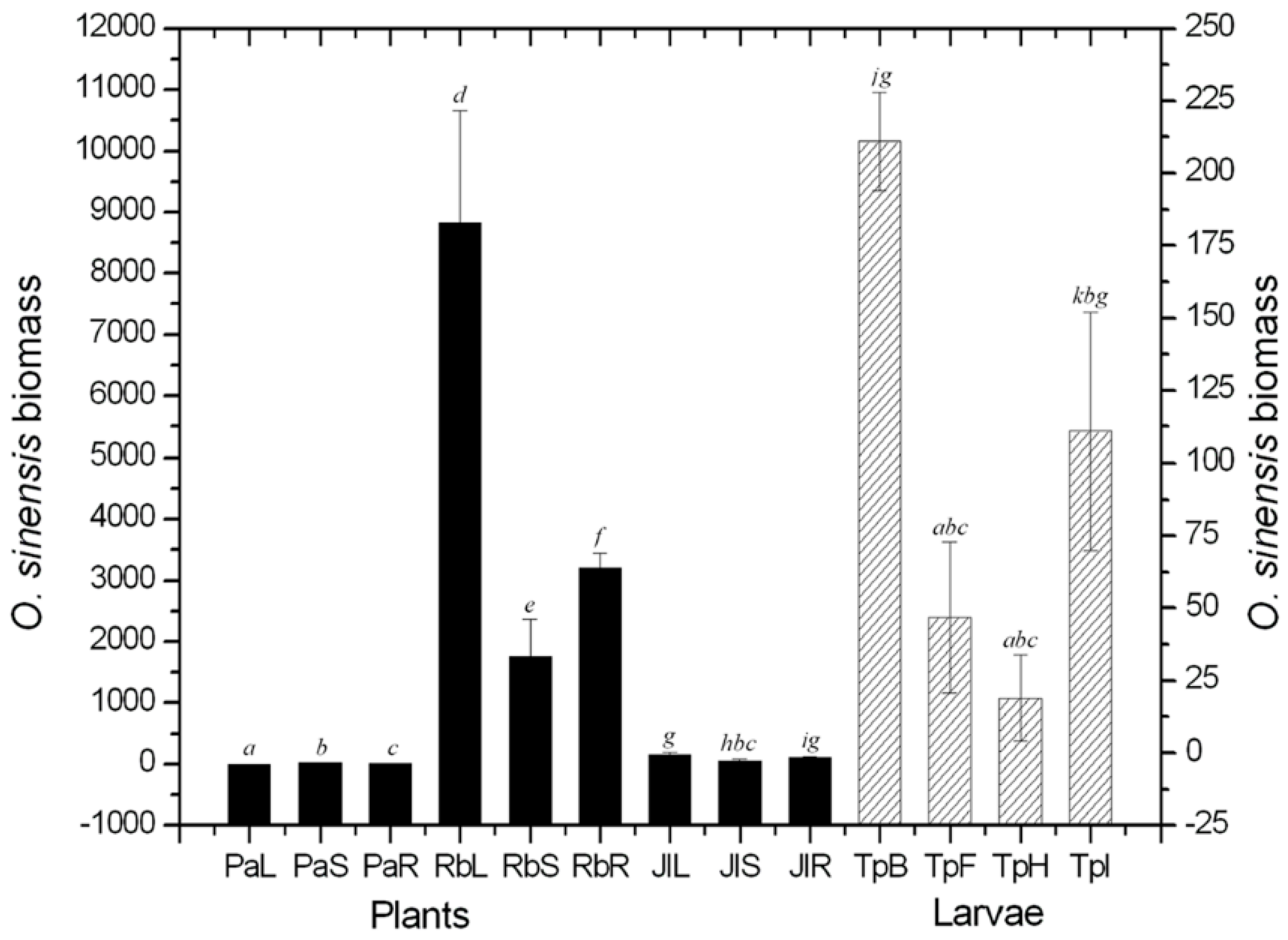
2.1. Quantificational Detection of O. sinensis Using Quantitative Polymerase Chain Reaction (qPCR)

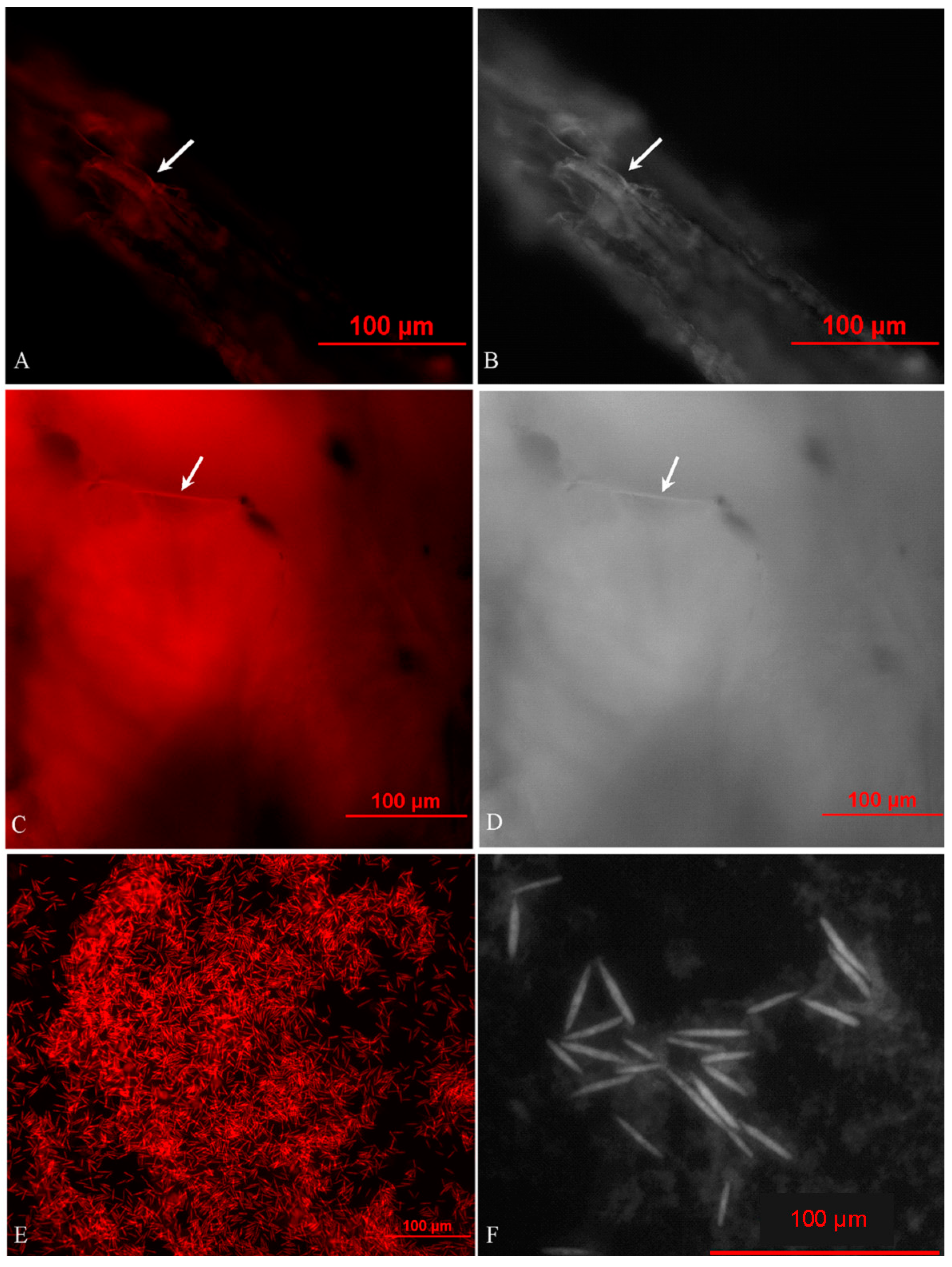
2.2. In Situ Investigation of O. sinensis Using Hybridization (FISH)
2.3. Effect of Culture with Plant Extracts on O. sinensis Mycelia
3. Discussion

4. Experimental Section
4.1. Materials
4.2. DNA Extraction and qPCR Assay
4.3. Hybridization (FISH) Protocol for in Situ Detection of O. sinensis
4.4. Effect of Plant Extracts on O. sinensis Mycelia
4.5. Data Analyses
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sung, G.-H.; Sung, J.-M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Winkler, D. Caterpillar fungus (Ophiocordyceps sinensis) production and sustainability on the Tibetan Plateau and in the Himalayas. Chin. J. Grassl. 2010, 32, 96–108. [Google Scholar] [CrossRef]
- Yoon, T.J.; Yu, K.W.; Shin, K.S.; Suh, H.J. Innate immune stimulation of exo-polymers prepared from Cordyceps sinensis by submerged culture. Appl. Microbiol. Biotechnol. 2008, 80, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Chioza, A.; Ohga, S. A review on fungal isolates reported as anamorphs of Ophiocordyceps sinensis. J. Mycol. 2014, 2014, 1–5. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Liu, X.Z.; Wang, M. Cloning, expression, and characterization of two novel cuticle-degrading serine proteases from the entomopathogenic fungus Cordyceps sinensis. Res. Microbiol. 2008, 159, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.W.; Liu, X.; Zhang, G.R. Revision of taxonomic system of the genus Hepialus (Lepidoptera, Hepialidae) currently adopted in China. J. Hunan Univ. Sci. Technol. 2010, 5, 114–120. [Google Scholar]
- Lei, W.; Chen, H.; Zhang, G.R.; Li, S.S.; Peng, Q.Y.; Zhong, X.; Liu, X. Molecular identification and food source inference of constructive plants, native to the Ophiocordyceps sinensis habitat. Afr. J. Biotechnol. 2011, 10, 159–167. [Google Scholar]
- Shrestha, G.; Zhang, W.M.; Zhang, Y.J.; Liu, X.Z. What is the Chinese caterpillar fungus Ophiocordyceps sinensis (Ophiocordycipitaceae)? Mycology 2010, 1, 228–236. [Google Scholar] [CrossRef]
- Nikoh, N.; Fukatsu, T. Interkingdom host jumping underground: Phylogenetic analysis of entomoparasitic fungi of the genus Cordyceps. Mol. Biol. Evol. 2000, 17, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Barseghyan, G.S.; Holliday, J.C.; Price, T.C.; Madison, L.M.; Wasser, S.P. Growth and cultural-morphological characteristics of vegetative mycelia of medicinal caterpillar fungus Ophiocordyceps sinensis G.H. Sung et al. (Ascomycetes) isolates from Tibetan Plateau (P.R. China). Int. J. Med. Mushrooms 2011, 13, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.C.; Hsieh, C.; Lin, F.Y.; Hsu, T.H. A systematic review of the mysterious caterpillar fungus Ophiocordyceps sinensis in Dong-ChongXiaCao (Dōng Chóng Xià Cǎo) and related bioactive ingredients. J. Tradit. Complement. Med. 2013, 3, 16–32. [Google Scholar] [PubMed]
- Norton, D.A.; Carpenter, M.A. Mistletoes as parasites: Host specificity and speciation. Trends Ecol. Evol. 1998, 13, 101–105. [Google Scholar] [CrossRef]
- Shaw, S.R. Euphorine phylogeny: The evolution of diversity in host-utilization by parasitoid wasps (Hymenoptera: Braconidae). Ecol. Entomol. 1998, 13, 323–335. [Google Scholar] [CrossRef]
- Hafner, M.S.; Nadler, S.A. Phylogenetic trees support the coevolution of parasites and their hosts. Nature 1988, 332, 258–259. [Google Scholar] [CrossRef] [PubMed]
- Maslov, D.A.; Lukes, J.; Jirku, M.; Simpson, L. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: Implication for the evolution of parasitism in the trypanosomatid protozoa. Mol. Biochem. Parasitol. 1996, 75, 197–205. [Google Scholar] [CrossRef]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Durette-Desset, M.C.; Beveridge, I.; Spratt, D.M. The origins and evolutionary expansion of the Strongylida (Nematoda). Int. J. Parasitol. 1994, 24, 1139–1165. [Google Scholar] [CrossRef]
- Millanes, A.M.; Truong, C.; Westberg, M.; Diederich, P.; Wedin, M. Host switching promotes diversity in host-specialized mycoparasitic fungi: Uncoupled evolution in the Biatoropsis-usnea system. Evolution 2014, 68, 1576–1593. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.S.; Wang, X.L.; Li, Y.; Wang, W.J.; Yang, R.H.; Ren, S.Y.; Yao, Y.J. Development of conventional and nested PCR assays for the detection of Ophiocordyceps sinensis. J. Basic Microb. 2013, 53, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Peng, Q.Y.; Li, S.S.; Chen, H.; Sun, H.X.; Zhang, G.R.; Liu, X. Detection of Ophiocordyceps sinensis in the roots of plants in alpine meadows by nested-touchdown polymerase chain reaction. Fungal Biol. 2014, 118, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Vasconcelos, V. Use of qPCR for the study of hepatotoxic cyanobacteria population dynamics. Arch Microbiol. 2011, 193, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Takai, K.; Horikoshi, K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 2000, 66, 5066–5072. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.J.; Osborn, A.M. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 2009, 67, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Maczey, N.; Dhendup, K.; Cannon, P.; Hywel-Jones, N.; Rai, T.B. Thitarodes namnai sp. nov. and T. Caligophilus sp. nov. (Lepidoptera: Hepialidae), hosts of the economically important entomopathogenic fungus Ophiocordyceps sinensis in Bhutan. Zootaxa 2010, 2412, 42–52. [Google Scholar]
- Zhang, G.R.; Yu, J.F.; Wu, G.G.; Liu, X. Factors influencing the occurrence of Ophiocordyceps sinensis. Acta Ecol. Sin. 2011, 31, 4117–4125. [Google Scholar]
- Hui, W.; Li, X.Y.; Zhang, Y.; Su, Z.C.; Zhang, C.G. Relationship between soil fungistasis and bacterial community structure. Chin. J. Appl. Ecol. 2008, 19, 1574–1578. [Google Scholar]
- Peng, Q.Y.; Zhong, X.; Lei, W.; Zhang, G.R.; Liu, X. Detection of Ophiocordyceps sinensis in soil by quantitative real-time PCR. Can. J. Microbiol. 2013, 59, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.Q.; Han, Y.F.; Liang, J.D.; Dong, X.; Du, W. Issues of concern in the studies of Ophiocordyceps sinensis. Microbiol. China 2010, 37, 1692–1697. [Google Scholar]
- Suh, S.-O.; Noda, H.; Blackwell, M. Insect symbiosis: Derivation of yeast-like endosymbionts within an entomopathogenic filamentous lineage. Mol. Biol. Evol. 2001, 18, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.X.; He, Y.H. The relationship and countermeasure of continuable utilize aweto resources in Tibet. Edible Fungal China 2007, 26, 18–20. [Google Scholar]
- Stone, R. Last stand for the body snatcher of the Himalayas? Science 2008, 322, 1182. [Google Scholar] [CrossRef] [PubMed]
- Cannon, P.F.; Hywel-Jones, N.L.; Maczey, N.; Norbu, L.; Tshitila; Samdup, T.; Lhendup, P. Steps towards sustainable harvest of Ophiocordyceps sinensis in Bhutan. Biodivers. Conserv. 2009, 18, 2263–2281. [Google Scholar] [CrossRef]
- Humber, R.A. Evolution of entomopathogenicity in fungi. J. Invertebr. Pathol. 2008, 98, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Agosta, S.J. On ecological fitting, plant-insect associations, herbivore host shifts, and host plant selection. Oikos 2006, 114, 556–565. [Google Scholar] [CrossRef]
- Tian, L.H.; Hu, B.; Zhou, H.; Zhang, W.M.; Qu, L.H.; Chen, Y.Q. Molecular phylogeny of the entomopathogenic fungi of the genus Cordyceps (Ascomycota: Clavicipitaceae) and its evolutionary implications. J. Syst. Evol. 2010, 48, 435–444. [Google Scholar] [CrossRef]
- Lei, W.; Li, S.S.; Peng, Q.Y.; Liu, X. A real-time qPCR assay to quantify Ophiocordyceps sinensis biomass in Thitarodes larvae. J. Microbiol. 2013, 51, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Krak, K.; Janouškov, P.C.; Vosátka, M.; Štorchová, H. Intraradical dynamics of two coexisting Isolates of the arbuscular mycorrhizal fungus Glomus intraradices sensu lato as estimated by real-time PCR of mitochondrial DNA. Appl. Environ. Microbiol. 2012, 78, 3630–3637. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X. Study on the Relationship between Ophiocordyceps sinensis and High Altitude Plants’ Roots. Ph.D. Thesis, Sun Yat-sen University, July 2010. [Google Scholar]
- Lei, W. Interdependent Relationship between Host Insects, Plants and Hirsutella sinensis in Alpine Habitat. Ph.D. Thesis, Sun Yat-sen University, July 2012. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, W.; Zhang, G.; Peng, Q.; Liu, X. Development of Ophiocordyceps sinensis through Plant-Mediated Interkingdom Host Colonization. Int. J. Mol. Sci. 2015, 16, 17482-17493. https://doi.org/10.3390/ijms160817482
Lei W, Zhang G, Peng Q, Liu X. Development of Ophiocordyceps sinensis through Plant-Mediated Interkingdom Host Colonization. International Journal of Molecular Sciences. 2015; 16(8):17482-17493. https://doi.org/10.3390/ijms160817482
Chicago/Turabian StyleLei, Wei, Guren Zhang, Qingyun Peng, and Xin Liu. 2015. "Development of Ophiocordyceps sinensis through Plant-Mediated Interkingdom Host Colonization" International Journal of Molecular Sciences 16, no. 8: 17482-17493. https://doi.org/10.3390/ijms160817482




