The miR-200 Family: Versatile Players in Epithelial Ovarian Cancer
Abstract
:1. Introduction
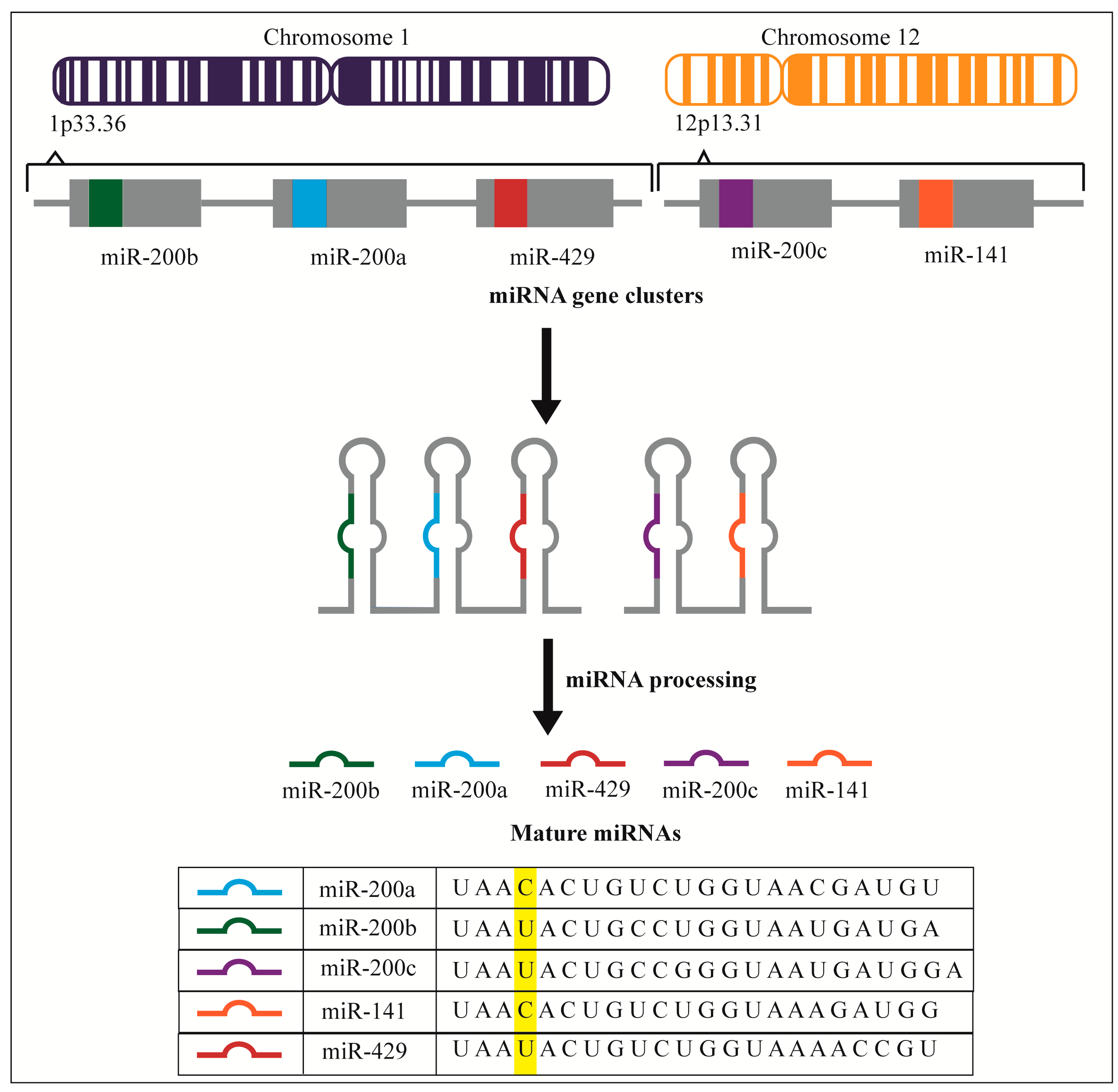
2. miR-200 Expression Profiles
| Study | Samples & Normal Controls | miRNA 200 Family Expression | Conclusions Made by Authors |
|---|---|---|---|
| Iorio et al. [44]; Ohio State Comprehensive Cancer Center microarray, version 2.0 with 460 mature miRNA probes (235 human miRNAs) | Samples: 69 malignant tumor tissues (including serous, endometrioid, clear cell, poorly differentiated and mucinous ovarian carcinoma); Controls: 15 normal ovarian tissue sections | Increased expression of miR-200a, 200b, 200c and 141 in tumor samples vs. normal tissue | MiR-200a, 200b, 200c, and 141 share a common putative target BAP1 (BRCA associated protein 1), a tumor suppressor down-regulated in ovarian cancer |
| Yang et al. [45]; Oligonucleotide array, GeneScreen Plus (NEN) membranes printed with 515 human and mouse miRNA probes | Samples: 10 human ovarian epithelial tumors; Controls: Normal ovarian tissue and immortalized human ovarian surface epithelium | 43% of primary ovarian carcinomas showed increased miR-200a expression | Increased miR-200a expression was associated with high grade and late stage disease |
| Dahiya et al. [46]; miRCURY™ LNA miRNA arrays with 1458 probes for all miRNAs in miRBase Release 8.1 (Exiqon) | Samples: 34 cancer tissues and 10 ovarian cancer cell lines (BG-1, UCI-101, HEY, OVCA420, OVCA432, OVCA433, OVCAR2, OVCAR3, OVCAR5, OV90); Controls: HOSE-B cells (human ovarian surface epithelial cell line immortalized with E6 and E7) | MiR-200a and 141 were found to be down-regulated in the neoplastic samples | Using Target Scan 3.0 miR-200a and 141 were found to share three predicted targets (ZEB2, KLF12 and ZFR) |
| Wyman et al. [47]; Parallel pyrosequencing (454 Life Sciences Platform) | Samples: Stage III/IV ovarian tumors including 19 serous, 4 clear cell and 10 endometrioid; Controls: 4 Normal primary human ovarian surface epithelium (HOSE) and E6/E7 immortalized HOSE | MiR-200a, 200b, 200c, 141, and 429 showed increased expression in ovarian tumors and the immortalized HOSE | Normal HOSE expresses low levels of miR-200 family. Immortalization induces their expression |
| Lee et al. [48]; Microarray with 668 Ambion probes (328 known and 154 predicted human miRNA probes) | Samples: 37 serous tumors (including high grade, low grade and borderline serous tumors); Controls: 3 normal fallopian tube epithelium sampled from the fimbriae | In high grade serous tumors miR-200c and 141 were up-regulated; In low grade serous tumors, miR-200a, 200b, 200c, and 141 were up-regulated | MiR-200a, 200b, 200c, and 141 were up-regulated in serous tumors. This was the first study that used fallopian tube epithelium as normal control as opposed to ovarian surface epithelium |
| Bendoraite et al. [49]; qRT-PCR using Taqman miRNA assays (Applied Biosciences) | Samples: Stage III/IV malignant ovarian primary tumors from 70 patients (including serous, endometrioid, and clear cell histotypes), 15 ovarian cancer cell lines (A1847, A2780, CaOV3, ES-2, HEY, IGROV1, OVCAR3, OVCAR5, OVCAR10, OV-90, PEO-1, SKOV3, TOV-21G, TOV-112D, 2008); Controls: Non-immortalized early passage primary cell cultures derived from HOSE as normal controls | Expression of all five members of miR-200 family were substantially higher in the primary tumors compared to normal tissues | Low expression of ZEB2 and high expression of miR-200 family in the tumor samples supports mesothelial to epithelial transition model |
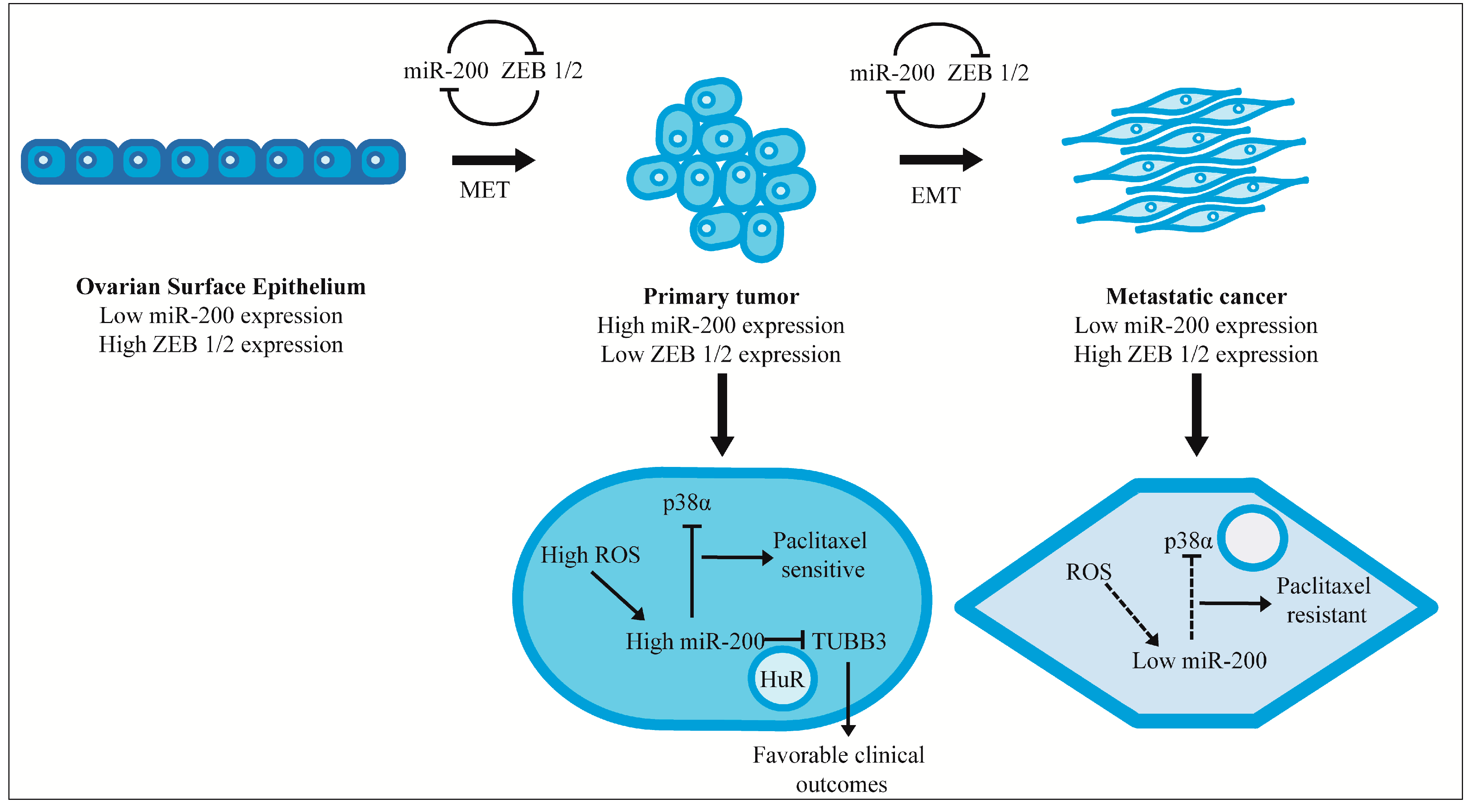
3. miR-200 and Metastasis
4. Effect on Chemotherapeutic Response and Clinical Outcomes
| Study | Samples | miRNA 200 Family Expression | Conclusions Made by Authors |
|---|---|---|---|
| Nam et al. [96]; Microarray with 377 (314 human) mirVana miRNA probes (Ambion) | Samples: 20 serous ovarian cancer tissues: 9 chemo-resistant, 11 chemo-sensitive tumors; Controls: 8 normal ovarian tissues | Increased expression of miR-200a, 200b, 200c and 141 in tumor samples vs. normal tissue | High expression of miR-200a, 200b, 200c and 141 were significantly correlated with decreased progression-free survival as well as overall survival |
| Hu et al. [97]; qRT-PCR miRNA assays (Applied Biosystems) | 55 patients: 48 epithelial ovarian carcinomas and 7 primary peritoneal carcinomas | Disease recurrence and poor overall survival were associated with low miR-200a, 200b and 429 expression | miR-200b-429 cluster expression has prognostic value in EOC |
| Eitan et al. [98]; Custom microarray slide (Nexetrion®) with 900 miRNA probes | 57 patients who had undergone surgery for tumor resection: 19 Stage I patients, 38 Stage III patients; All received platinum based chemotherapy | miR-200a expression was higher in Stage I ovarian cancer compared to Stage II | The data set shows significantly higher expression of miR-200a in early stage disease correlating with improved survival |
| Marchini et al. [89]; G4470B human miRNA microarray (Agilent Technologies) with probes for 723 human miRNAs | 144 patients with Stage I EOC out of which 29 patients relapsed | Tumors with lower miR-200c levels seen in patients who relapsed | miR-200 expression could be used as an indication of relapse in Stage I tumors |
| Leskela et al. [88]; qRT-PCR using the miRCURY™ LNA miRNA assay kits (Exiqon) | 72 patients were studied for overall survival analysis; A subgroup of 57 patients with both advanced tumor stage and serous carcinoma histotype were studied for treatment response | miR-200 expression correlated with β-Tubulin III levels | Low miR-200 expression was seen in patients without complete response to paclitaxel when compared to patients with complete response; Low miR-200 expression had a trend towards poor survival |
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–73. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Jaskiewicz, L.; Kolb, F.A.; Pillai, R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005, 15, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Hausser, J.; Syed, A.P.; Bilen, B.; Zavolan, M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013, 23, 604–615. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Visone, R.; Croce, C.M. MiRNAs and cancer. Am. J. Pathol. 2009, 174, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Lieberman, J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal. 2015, 8, re3. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Dolan, M.E. The emerging role of microRNAs in drug responses. Curr. Opin. Mol. Ther. 2010, 12, 695–702. [Google Scholar] [PubMed]
- Chan, E.; Prado, D.E.; Weidhaas, J.B. Cancer microRNAs: From subtype profiling to predictors of response to therapy. Trends Mol. Med. 2011, 17, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Bar-Eli, M. Searching for the “melano-miRs”: miR-214 drives melanoma metastasis. EMBO J. 2011, 30, 1880–1881. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013, 774, 1–20. [Google Scholar] [PubMed]
- Mezzanzanica, D.; Bagnoli, M.; de Cecco, L.; Valeri, B.; Canevari, S. Role of microRNAs in ovarian cancer pathogenesis and potential clinical implications. Int. J. Biochem. Cell Biol. 2010, 42, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Welsh, J.W.; Calin, G.A. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012, 195, 151–161. [Google Scholar] [PubMed]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug. Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.Y.; Calin, G.A. MicroRNAs as therapeutic targets in human cancers. Wiley Interdiscip. Rev. RNA 2014, 5, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.J.; Bahal, R.; Babar, I.A.; Pincus, Z.; Barrera, F.; Liu, C.; Svoronos, A.; Braddock, D.T.; Glazer, P.M.; Engelman, D.M.; et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 2015, 518, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Redis, R.S.; Calin, S.; Yang, Y.; You, M.J.; Calin, G.A. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol. Ther. 2012, 136, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Kohlhapp, F.J.; Mitra, A.K.; Lengyel, E.; Peter, M.E. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, N.; Morin, P.J. MicroRNAs in ovarian carcinomas. Endocr. Relat. Cancer 2010, 17, F77–F89. [Google Scholar] [CrossRef] [PubMed]
- Van Jaarsveld, M.T.; Helleman, J.; Berns, E.M.; Wiemer, E.A. MicroRNAs in ovarian cancer biology and therapy resistance. Int. J. Biochem. Cell Biol. 2010, 42, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Croce, C.M. The role of microRNAs in the tumorigenesis of ovarian cancer. Front. Oncol. 2013, 3, 153. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kim, S.; Kim, I.M. Regulation of metastasis by microRNAs in ovarian cancer. Front. Oncol. 2014, 4, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lu, Z.; Unruh, A.K.; Ivan, C.; Baggerly, K.A.; Calin, G.A.; Li, Z.; Bast, R.C.; Le, X.F. Clinically relevant microRNAs in ovarian cancer. Mol. Cancer Res. 2015, 13, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C.; Hennessy, B.; Mills, G.B. The biology of ovarian cancer: New opportunities for translation. Nat. Rev. Cancer 2009, 9, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Köbel, M.; Kalloger, S.E.; Boyd, N.; McKinney, S.; Mehl, E.; Palmer, C.; Leung, S.; Bowen, N.J.; Ionescu, D.N.; Rajput, A.; et al. Ovarian carcinoma subtypes are different diseases: Implications for biomarker studies. PLoS Med. 2008, 5, e232. [Google Scholar] [CrossRef] [PubMed]
- Roett, M.A.; Evans, P. Ovarian cancer: An overview. Am. Fam. Physician 2009, 80, 609–616. [Google Scholar] [PubMed]
- Marchetti, C.; Pisano, C.; Facchini, G.; Bruni, G.S.; Magazzino, F.P.; Losito, S.; Pignata, S. First-line treatment of advanced ovarian cancer: Current research and perspectives. Expert Rev. Anticancer Ther. 2010, 10, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Volinia, S.; Bonome, T.; Calin, G.A.; Greshock, J.; Yang, N.; Liu, C.G.; Giannakakis, A.; Alexiou, P.; Hasegawa, K.; et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl. Acad. Sci. USA. [CrossRef] [PubMed]
- Yang, D.; Sun, Y.; Hu, L.; Zheng, H.; Ji, P.; Pecot, C.V.; Zhao, Y.; Reynolds, S.; Cheng, H.; Rupaimoole, R.; et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell 2013, 23, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Yang, C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [PubMed]
- Zaravinos, A. The regulatory role of microRNAs in EMT and cancer. J. Oncol. 2015, 2015, 865816. [Google Scholar] [CrossRef] [PubMed]
- Gadducci, A.; Sergiampietri, C.; Lanfredini, N.; Guiggi, I. Micro-RNAs and ovarian cancer: The state of art and perspectives of clinical research. Gynecol. Endocrinol. 2014, 30, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Kong, W.; He, L.; Zhao, J.J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, N.; Sherman-Baust, C.A.; Wang, T.L.; Davidson, B.; Shih, I.M.; Zhang, Y.; Wood, W.; Becker, K.G.; Morin, P.J. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE 2008, 3, e2436. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Parkin, R.K.; Mitchell, P.S.; Fritz, B.R.; O’Briant, K.; Godwin, A.K.; Urban, N.; Drescher, C.W.; Knudsen, B.S.; Tewari, M. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE 2009, 4, e5311. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Subramanian, S.; Beck, A.H.; Espinosa, I.; Senz, J.; Zhu, S.X.; Huntsman, D.; van de Rijn, M.; Gilks, C.B. MicroRNA profiling of BRCA1/2 mutation-carrying and non-mutation-carrying high-grade serous carcinomas of ovary. PLoS ONE 2009, 4, e7314. [Google Scholar] [CrossRef] [PubMed]
- Bendoraite, A.; Knouf, E.C.; Garg, K.S.; Parkin, R.K.; Kroh, E.M.; O’Briant, K.C.; Ventura, A.P.; Godwin, A.K.; Karlan, B.Y.; Drescher, C.W.; et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: Evidence supporting a mesothelial-to-epithelial transition. Gynecol. Oncol. 2010, 116, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Erickson, B.K.; Conner, M.G.; Landen, C.N. The role of the fallopian tube in the origin of ovarian cancer. Am. J. Obstet. Gynecol. 2013, 209, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Dubeau, L.; Drapkin, R. Coming into focus: The nonovarian origins of ovarian cancer. Ann. Oncol. 2013, 24, viii28–viii35. [Google Scholar] [CrossRef] [PubMed]
- Crum, C.P.; Herfs, M.; Ning, G.; Bijron, J.G.; Howitt, B.E.; Jimenez, C.A.; Hanamornroongruang, S.; McKeon, F.D.; Xian, W. Through the glass darkly: Intraepithelial neoplasia, top-down differentiation, and the road to ovarian cancer. J. Pathol. 2013, 231, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Auersperg, N. Ovarian surface epithelium as a source of ovarian cancers: Unwarranted speculation or evidence-based hypothesis? Gynecol. Oncol. 2013, 130, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Zorn, K.K.; Jazaeri, A.A.; Awtrey, C.S.; Gardner, G.J.; Mok, S.C.; Boyd, J.; Birrer, M.J. Choice of normal ovarian control influences determination of differentially expressed genes in ovarian cancer expression profiling studies. Clin. Cancer Res. 2003, 9, 4811–4818. [Google Scholar] [PubMed]
- Merritt, M.A.; Bentink, S.; Schwede, M.; Iwanicki, M.P.; Quackenbush, J.; Woo, T.; Agoston, E.S.; Reinhardt, F.; Crum, C.P.; Berkowitz, R.S.; et al. Gene expression signature of normal cell-of-origin predicts ovarian tumor outcomes. PLoS ONE 2013, 8, e80314. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta 2012, 1826, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kan, C.W.; Hahn, M.A.; Gard, G.B.; Maidens, J.; Huh, J.Y.; Marsh, D.J.; Howell, V.M. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer 2012, 12, 627. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, S.; Zhang, J.D.; Schwäger, A.; Mannsperger, H.; Riazalhosseini, Y.; Burmester, S.; Ward, A.; Korf, U.; Wiemann, S.; Sahin, O. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 2010, 29, 4297–4306. [Google Scholar] [CrossRef] [PubMed]
- Christoffersen, N.R.; Silahtaroglu, A.; Orom, U.A.; Kauppinen, S.; Lund, A.H. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA 2007, 13, 1172–1178. [Google Scholar] [CrossRef] [PubMed]
- Hurteau, G.J.; Carlson, J.A.; Spivack, S.D.; Brock, G.J. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007, 67, 7972–7976. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910–14914. [Google Scholar] [CrossRef] [PubMed]
- Comijn, J.; Berx, G.; Vermassen, P.; Verschueren, K.; van Grunsven, L.; Bruyneel, E.; Mareel, M.; Huylebroeck, D.; van Roy, F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 2001, 7, 1267–1278. [Google Scholar] [CrossRef]
- Vandewalle, C.; Comijn, J.; de Craene, B.; Vermassen, P.; Bruyneel, E.; Andersen, H.; Tulchinsky, E.; van Roy, F.; Berx, G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005, 33, 6566–6578. [Google Scholar] [CrossRef] [PubMed]
- Eger, A.; Aigner, K.; Sonderegger, S.; Dampier, B.; Oehler, S.; Schreiber, M.; Berx, G.; Cano, A.; Beug, H.; Foisner, R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005, 24, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Aigner, K.; Dampier, B.; Descovich, L.; Mikula, M.; Sultan, A.; Schreiber, M.; Mikulits, W.; Brabletz, T.; Strand, D.; Obrist, P.; et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 2007, 26, 6979–6988. [Google Scholar] [CrossRef] [PubMed]
- Vergara, D.; Merlot, B.; Lucot, J.P.; Collinet, P.; Vinatier, D.; Fournier, I.; Salzet, M. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett. 2010, 291, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Kang, Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008, 5, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bracken, C.P.; Bert, A.G.; Goodall, G.J. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 2008, 7, 3112–3118. [Google Scholar] [CrossRef] [PubMed]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Brabletz, T. The ZEB/miR-200 feedback loop—A motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, N.; Reavis, A.N.; McDonald, J.F. Sequence variation among members of the miR-200 microRNA family is correlated with variation in the ability to induce hallmarks of mesenchymal-epithelial transition in ovarian cancer cells. J. Ovarian Res. 2014, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, K.; Kajiyama, H.; Shibata, K.; Yuan, H.; Kikkawa, F.; Senga, T. Expression of the miR200 family of microRNAs in mesothelial cells suppresses the dissemination of ovarian cancer cells. Mol. Cancer Ther. 2014, 13, 2081–2091. [Google Scholar] [CrossRef] [PubMed]
- Pecot, C.V.; Rupaimoole, R.; Yang, D.; Akbani, R.; Ivan, C.; Lu, C.; Wu, S.; Han, H.D.; Shah, M.Y.; Rodriguez-Aguayo, C.; et al. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013, 4, 2427. [Google Scholar] [CrossRef] [PubMed]
- Zaman, M.S.; Maher, D.M.; Khan, S.; Jaggi, M.; Chauhan, S.C. Current status and implications of microRNAs in ovarian cancer diagnosis and therapy. J. Ovarian Res. 2012, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Scomparin, A.; Polyak, D.; Krivitsky, A.; Satchi-Fainaro, R. Achieving successful delivery of oligonucleotides—From physico-chemical characterization to in vivo evaluation. Biotechnol. Adv. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef] [PubMed]
- Koutsaki, M.; Spandidos, D.A.; Zaravinos, A. Epithelial-mesenchymal transition-associated miRNAs in ovarian carcinoma, with highlight on the miR-200 family: Prognostic value and prospective role in ovarian cancer therapeutics. Cancer Lett. 2014, 351, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Mozzetti, S.; Ferlini, C.; Concolino, P.; Filippetti, F.; Raspaglio, G.; Prislei, S.; Gallo, D.; Martinelli, E.; Ranelletti, F.O.; Ferrandina, G.; et al. Class III beta-tubulin overexpression is a prominent mechanism of paclitaxel resistance in ovarian cancer patients. Clin. Cancer Res. 2005, 11, 298–305. [Google Scholar] [PubMed]
- Kavallaris, M.; Annereau, J.P.; Barret, J.M. Potential mechanisms of resistance to microtubule inhibitors. Semin. Oncol. 2008, 35, S22–S27. [Google Scholar] [CrossRef] [PubMed]
- Umezu, T.; Shibata, K.; Kajiyama, H.; Terauchi, M.; Ino, K.; Nawa, A.; Kikkawa, F. Taxol resistance among the different histological subtypes of ovarian cancer may be associated with the expression of class III β-tubulin. Int. J. Gynecol. Pathol. 2008, 27, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, D.R.; Spoelstra, N.S.; Howe, E.N.; Nordeen, S.K.; Richer, J.K. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer Ther. 2009, 8, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, D.R.; Howe, E.N.; Spoelstra, N.S.; Richer, J.K. Loss of miR-200c: A marker of aggressiveness and chemoresistance in female reproductive Cancers. J. Oncol. 2010, 2010, 821717. [Google Scholar] [CrossRef] [PubMed]
- Leskelä, S.; Leandro-García, L.J.; Mendiola, M.; Barriuso, J.; Inglada-Pérez, L.; Muñoz, I.; Martínez-Delgado, B.; Redondo, A.; de Santiago, J.; Robledo, M.; et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr. Relat. Cancer 2011, 18, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Marchini, S.; Cavalieri, D.; Fruscio, R.; Calura, E.; Garavaglia, D.; Fuso Nerini, I.; Mangioni, C.; Cattoretti, G.; Clivio, L.; Beltrame, L.; et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: A retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011, 12, 273–285. [Google Scholar] [CrossRef]
- Raspaglio, G.; de Maria, I.; Filippetti, F.; Martinelli, E.; Zannoni, G.F.; Prislei, S.; Ferrandina, G.; Shahabi, S.; Scambia, G.; Ferlini, C. HuR regulates beta-tubulin isotype expression in ovarian cancer. Cancer Res. 2010, 70, 5891–5900. [Google Scholar] [CrossRef] [PubMed]
- Prislei, S.; Martinelli, E.; Mariani, M.; Raspaglio, G.; Sieber, S.; Ferrandina, G.; Shahabi, S.; Scambia, G.; Ferlini, C. MiR-200c and HuR in ovarian cancer. BMC Cancer 2013, 13, 72. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Lu, K.; Dai, S.; Hu, Y.; Fan, W. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 2392–2401. [Google Scholar] [PubMed]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; de Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.P.; Cottu, P.; Sastre-Garau, X.; et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, B.; Jan, K.Y.; Chen, C.H.; Hour, T.C.; Yu, H.J.; Pu, Y.S. Resistance to paclitaxel is proportional to cellular total antioxidant capacity. Cancer Res. 2005, 65, 8455–8460. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, J.; Hu, Y.; Lu, W.; Pelicano, H.; Huang, P. Novel action of paclitaxel against cancer cells: Bystander effect mediated by reactive oxygen species. Cancer Res. 2007, 67, 3512–3517. [Google Scholar] [CrossRef] [PubMed]
- Nam, E.J.; Yoon, H.; Kim, S.W.; Kim, H.; Kim, Y.T.; Kim, J.H.; Kim, J.W.; Kim, S. MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 2008, 14, 2690–2695. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Macdonald, D.M.; Huettner, P.C.; Feng, Z.; el Naqa, I.M.; Schwarz, J.K.; Mutch, D.G.; Grigsby, P.W.; Powell, S.N.; Wang, X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol. Oncol. 2009, 114, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Eitan, R.; Kushnir, M.; Lithwick-Yanai, G.; David, M.B.; Hoshen, M.; Glezerman, M.; Hod, M.; Sabah, G.; Rosenwald, S.; Levavi, H. Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecol. Oncol. 2009, 114, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Vilming Elgaaen, B.; Olstad, O.K.; Haug, K.B.; Brusletto, B.; Sandvik, L.; Staff, A.C.; Gautvik, K.M.; Davidson, B. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer 2014, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.C.; Wu, J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muralidhar, G.G.; Barbolina, M.V. The miR-200 Family: Versatile Players in Epithelial Ovarian Cancer. Int. J. Mol. Sci. 2015, 16, 16833-16847. https://doi.org/10.3390/ijms160816833
Muralidhar GG, Barbolina MV. The miR-200 Family: Versatile Players in Epithelial Ovarian Cancer. International Journal of Molecular Sciences. 2015; 16(8):16833-16847. https://doi.org/10.3390/ijms160816833
Chicago/Turabian StyleMuralidhar, Goda G., and Maria V. Barbolina. 2015. "The miR-200 Family: Versatile Players in Epithelial Ovarian Cancer" International Journal of Molecular Sciences 16, no. 8: 16833-16847. https://doi.org/10.3390/ijms160816833





