Get to Understand More from Single-Cells: Current Studies of Microfluidic-Based Techniques for Single-Cell Analysis
Abstract
:1. Introduction
2. Isolation, Trapping and Manipulation of Single Cells
| Approaches | Main Applications | Major Advantage | Major Disadvantage |
|---|---|---|---|
| Droplet-based microfluidics |
| High throughput screening for specific single-cells | Challenge to encapsulate single-cells in each droplet |
| |||
| Hydrodynamic trap |
| Multifunction in one device | Complicated fabricating process |
| |||
| Magnetic trap |
| Efficient trapping of labeled cells | Requires antibodies or primers for magnetic label |
| Acoustic trap |
| Good for cell positioning | May have negative effect to cells |
| Dielectrophoretic trap |
| Easily select target cells via alternating frequency of AC | Heat problem during long-term manipulation |
| |||
| Optical trap |
| Applicable in many fields of study | Requires expensive optical system |
|
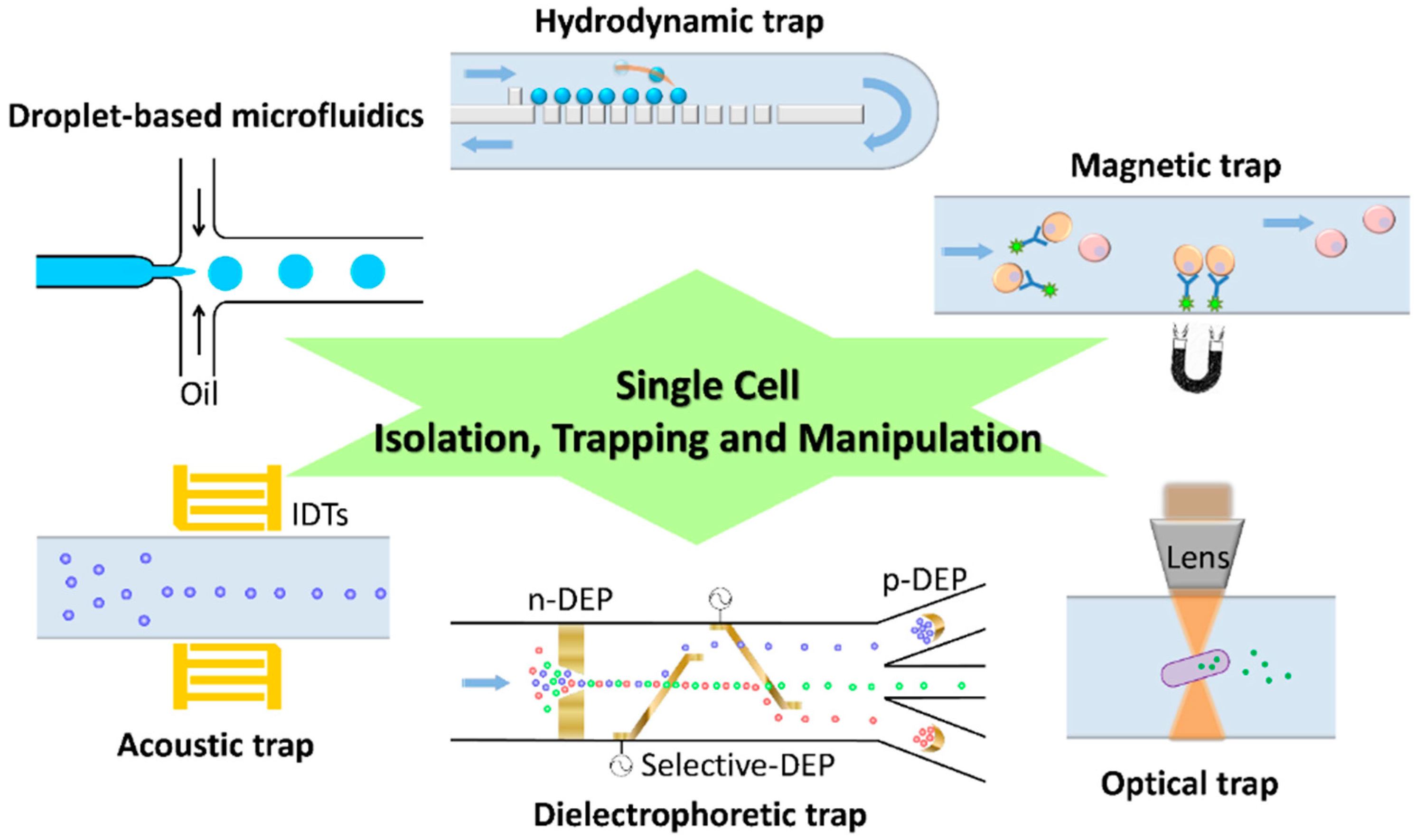
2.1. Hydrodynamic Trap
2.2. Optical Trap
2.3. Magnetic Trap
2.4. Dielectrophoretic Trap
2.5. Acoustic Trap
3. Single-Cell Analysis in a Microfluidic Device
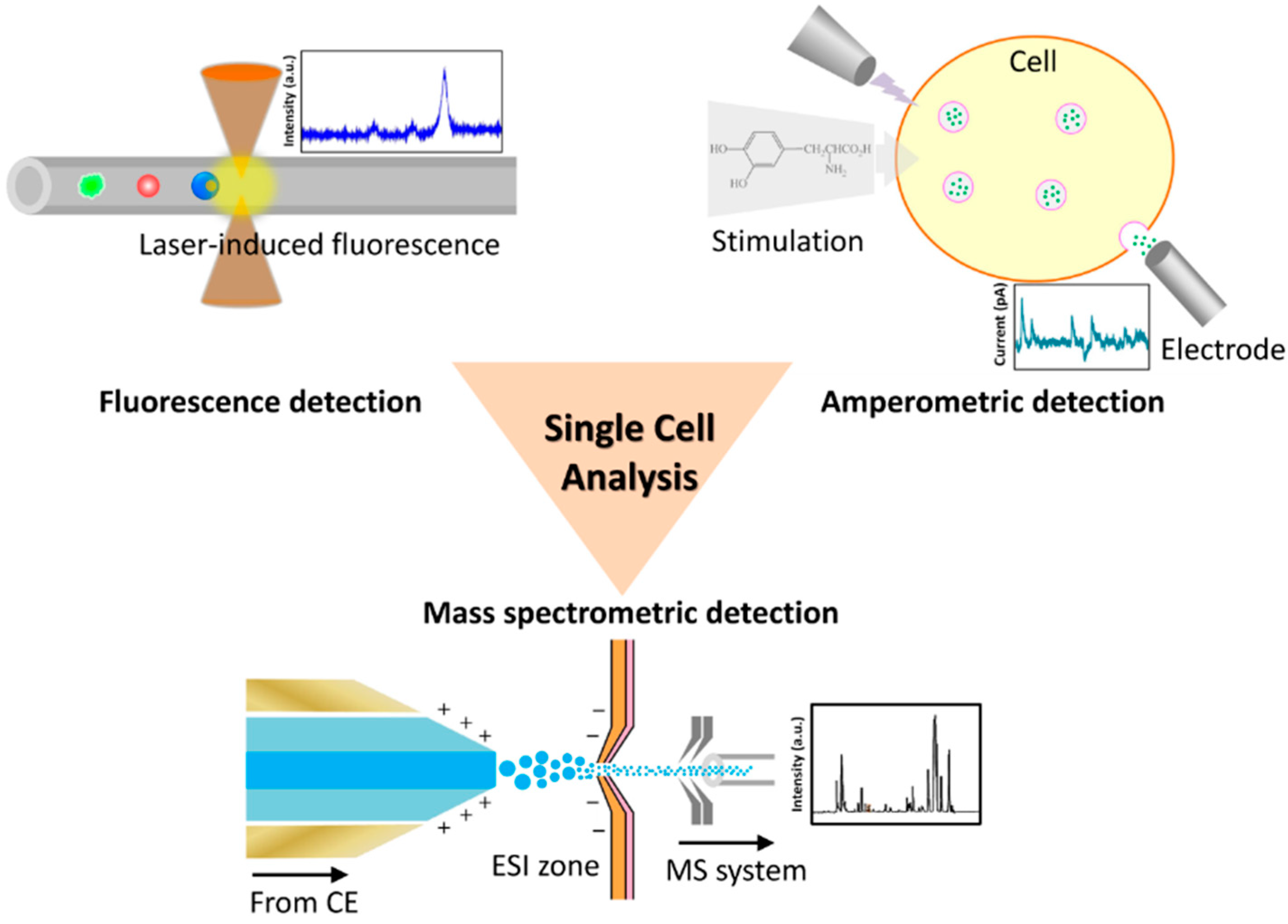
3.1. Fluorescence Detection
3.2. Amperometric Detection
3.3. Mass Spectrometric Detection
4. Application Summary of Single-Cell Analysis
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Junker, J.P.; van Oudenaarden, A. Every cell is special: Genome-wide studies add a new dimension to single-cell biology. Cell 2014, 157, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.B.; Marshall, D. Microfluidics for single cell analysis. Curr. Opin. Biotechnol. 2012, 23, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.N.; Zhou, J.H.; Wu, H.K. Materials for microfluidic chip fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef] [PubMed]
- Villar, G.; Graham, A.D.; Bayley, H. A tissue-like printed material. Science 2013, 340, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Chen, Y.A.; Liu, Y.J.; Yao, D.J. A multilayer concentric filter device to diminish clogging for separation of particles and microalgae based on size. Lab Chip 2014, 14, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Mosadegh, B.; Kuo, C.H.; Tung, Y.C.; Torisawa, Y.S.; Bersano-Begey, T.; Tavana, H.; Takayama, S. Integrated elastomeric components for autonomous regulation of sequential and oscillatory flow switching in microfluidic devices. Nat. Phys. 2010, 6, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Godin, J.M.; Chen, C.H.; Qiao, W.; Lee, H.; Lo, Y.H. Review Article: Recent advancements in optofluidic flow cytometer. Biomicrofluidics 2010, 4, 43001. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, W.J.; Pappas, D. Recent advances in microfluidic cell separations. Analyst 2013, 138, 4714–4721. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.L.; Ghaderi, A.; Zhou, H.; Agresti, J.; Weitz, D.A.; Fink, G.R.; Stephanopoulos, G. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption. Nat. Biotechnol. 2014, 32, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Nugent, E.; Javer, A.; Cicuta, P.; Sclavi, B.; Lagomarsino, M.C.; Dorfman, K.D. Microfluidic chemostat for measuring single cell dynamics in bacteria. Lab Chip 2013, 13, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.Y.; Chu, Y.S.; Thiery, J.P.; Lim, C.T.; Rodriguez, I. Microfluidic cell trap array for controlled positioning of single cells on adhesive micropatterns. Lab Chip 2013, 13, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Sochol, R.D.; Dueck, M.E.; Li, S.; Lee, L.P.; Lin, L.W. Hydrodynamic resettability for a microfluidic particulate-based arraying system. Lab Chip 2012, 12, 5051–5056. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, X.L.; Feng, X.J.; Liu, C.; Chen, P.; Chen, D.J.; Liu, B.F. A microfluidic digital single-cell assay for the evaluation of anticancer drugs. Anal. Bioanal. Chem. 2015, 407, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Ramser, K.; Hanstorp, D. Optical manipulation for single-cell studies. J. Biophotonics 2010, 3, 187–206. [Google Scholar] [CrossRef] [PubMed]
- Liberale, C.; Cojoc, G.; Bragheri, F.; Minzioni, P.; Perozziello, G.; la Rocca, R.; Ferrara, L.; Rajamanickam, V.; di Fabrizio, E.; Cristiani, I. Integrated microfluidic device for single-cell trapping and spectroscopy. Sci. Rep. 2013, 3, 1258. [Google Scholar] [CrossRef] [PubMed]
- Zehtabi-Oskuie, A.; Jiang, H.; Cyr, B.R.; Rennehan, D.W.; Al-Balushi, A.A.; Gordon, R. Double nanohole optical trapping: Dynamics and protein-antibody co-trapping. Lab Chip 2013, 13, 2563–2568. [Google Scholar] [CrossRef] [PubMed]
- Kotnala, A.; Gordon, R. Double nanohole optical tweezers visualize protein p53 suppressing unzipping of single DNA-hairpins. Biomed. Opt. Express 2014, 5, 1886–1894. [Google Scholar] [CrossRef] [PubMed]
- Kotnala, A.; DePaoli, D.; Gordon, R. Sensing nanoparticles using a double nanohole optical trap. Lab Chip 2013, 13, 4142–4146. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.C.; Wang, C.H.; Liou, T.M.; Lee, G.B. Influenza A virus-specific aptamers screened by using an integrated microfluidic system. Lab Chip 2014, 14, 2002–2013. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Byvank, T.; Chang, W.J.; Bharde, A.; Vieira, G.; Miller, B.L.; Chalmers, J.J.; Bashir, R.; Sooryakumar, R. On-chip magnetic separation and encapsulation of cells in droplets. Lab Chip 2013, 13, 1172–1181. [Google Scholar] [CrossRef] [PubMed]
- Nawarathna, D.; Norouzi, N.; McLane, J.; Sharma, H.; Sharac, N.; Grant, T.; Chen, A.; Strayer, S.; Ragan, R.; Khine, M. Shrink-induced sorting using integrated nanoscale magnetic traps. Appl. Phys. Lett. 2013, 102, 63504. [Google Scholar] [CrossRef] [PubMed]
- Jubery, T.Z.; Srivastava, S.K.; Dutta, P. Dielectrophoretic separation of bioparticles in microdevices: A review. Electrophoresis 2014, 35, 691–713. [Google Scholar] [CrossRef] [PubMed]
- Gossett, D.R.; Weaver, W.M.; Mach, A.J.; Hur, S.C.; Tse, H.T.K.; Lee, W.; Amini, H.; di Carlo, D. Label-free cell separation and sorting in microfluidic systems. Anal. Bioanal. Chem. 2010, 397, 3249–3267. [Google Scholar] [CrossRef] [PubMed]
- Taff, B.M.; Voldman, J. A scalable addressable positive-dielectrophoretic cell-sorting array. Anal. Chem. 2005, 77, 7976–7983. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Chao, T.C.; Ariyasinghe, N.; Ruiz, Y.; Lake, D.; Ros, R.; Ros, A. Selective trapping of single mammalian breast cancer cells by insulator-based dielectrophoresis. Anal. Bioanal. Chem. 2014, 406, 1855–1865. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.; Liu, C.X.; Stakenborg, J.L.T.; Lagae, L. Single cell viability observation in cell dielectrophoretic trapping on a microchip. Appl. Phys. Lett. 2014, 104, 013703. [Google Scholar] [CrossRef]
- Martinez-Duarte, R.; Camacho-Alanis, F.; Renaud, P.; Ros, A. Dielectrophoresis of lambda-DNA using 3D carbon electrodes. Electrophoresis 2013, 34, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.J.; Ahmed, D.; Mao, X.; Lin, S.C.S.; Lawit, A.; Huang, T.J. Acoustic tweezers: Patterning cells and microparticles using standing surface acoustic waves (SSAW). Lab Chip 2009, 9, 2890–2895. [Google Scholar] [CrossRef] [PubMed]
- Evander, M.; Johansson, L.; Lilliehorn, T.; Piskur, J.; Lindvall, M.; Johansson, S.; Almqvist, M.; Laurell, T.; Nilsson, J. Noninvasive acoustic cell trapping in a microfluidic perfusion system for online bioassays. Anal. Chem. 2007, 79, 2984–2991. [Google Scholar] [CrossRef] [PubMed]
- Hultstrom, J.; Manneberg, O.; Dopf, K.; Hertz, H.M.; Brismar, H.; Wiklund, M. Proliferation and viability of adherent cells manipulated by standing-wave ultrasound in a microfluidic chip. Ultrasound Med. Biol. 2007, 33, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, S.; Gu, Y.; Li, P.; Ding, X.; Wang, L.; McCoy, J.P.; Levine, S.J.; Huang, T.J. Continuous enrichment of low-abundance cell samples using standing surface acoustic waves (SSAW). Lab Chip 2014, 14, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.L.; Zhao, C.L.; Zhao, Y.H.; Li, S.X.; Rufo, J.; Yang, S.K.; Guo, F.; Huang, T.J. Optoacoustic tweezers: A programmable, localized cell concentrator based on opto-thermally generated, acoustically activated, surface bubbles. Lab Chip 2013, 13, 1772–1779. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Q.; Feng, J.; Tong, L.L.; Tang, B. Highly sensitive and homogeneous detection of membrane protein on a single living cell by aptamer and nicking enzyme assisted signal amplification based on microfluidic droplets. Anal. Chem. 2014, 86, 5101–5107. [Google Scholar] [CrossRef] [PubMed]
- Keithley, R.B.; Rosenthal, A.S.; Essaka, D.C.; Tanaka, H.; Yoshimura, Y.; Palcic, M.M.; Hindsgaul, O.; Dovichi, N.J. Capillary electrophoresis with three-color fluorescence detection for the analysis of glycosphingolipid metabolism. Analyst 2013, 138, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Metto, E.C.; Evans, K.; Barney, P.; Culbertson, A.H.; Gunasekara, D.B.; Caruso, G.; Huvey, M.K.; da Silva, J.A.F.; Lunte, S.M.; Culbertson, C.T. An integrated microfluidic device for monitoring changes in nitric oxide production in single T-lymphocyte (jurkat) cells. Anal. Chem. 2013, 85, 10188–10195. [Google Scholar] [CrossRef] [PubMed]
- Ban, E.; Chae, D.K.; Song, E.J. Determination of micro-RNA in cardiomyoblast cells using CE with LIF detection. Electrophoresis 2013, 34, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Ban, E.; Chae, D.K.; Song, E.J. Simultaneous detection of multiple microRNAs for expression profiles of microRNAs in lung cancer cell lines by capillary electrophoresis with dual laser-induced fluorescence. J. Chromatogr. A 2013, 1315, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Omiatek, D.M.; Cans, A.S.; Heien, M.L.; Ewing, A.G. Analytical approaches to investigate transmitter content and release from single secretory vesicles. Anal. Bioanal. Chem. 2010, 397, 3269–3279. [Google Scholar] [CrossRef] [PubMed]
- Omiatek, D.M.; Dong, Y.; Heien, M.L.; Ewing, A.G. Only a fraction of quantal content is released during exocytosis as revealed by electrochemical cytometry of secretory vesicles. ACS Chem. Neurosci. 2010, 1, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Larsen, S.T.; Heien, M.L.; Taboryski, R. Amperometric noise at thin film band electrodes. Anal. Chem. 2012, 84, 7744–7749. [Google Scholar] [CrossRef] [PubMed]
- Larsen, S.T.; Vreeland, R.F.; Heien, M.L.; Taboryski, R. Characterization of poly(3,4-ethylenedioxythiophene):tosylate conductive polymer microelectrodes for transmitter detection. Analyst 2012, 137, 1831–1836. [Google Scholar] [CrossRef] [PubMed]
- Kleparnik, K. Recent advances in the combination of capillary electrophoresis with mass spectrometry: From element to single-cell analysis. Electrophoresis 2013, 34, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Liu, H.X.; Jiang, Y.Y.; Lin, J.M. Recent advances in microfluidics combined with mass spectrometry: Technologies and applications. Lab Chip 2013, 13, 3309–3322. [Google Scholar] [CrossRef] [PubMed]
- Aerts, J.T.; Louis, K.R.; Crandall, S.R.; Govindaiah, G.; Cox, C.L.; Sweedler, J.V. Patch clamp electrophysiology and capillary electrophoresis-mass spectrometry metabolomics for single cell characterization. Anal. Chem. 2014, 86, 3203–3208. [Google Scholar] [CrossRef] [PubMed]
- Mellors, J.S.; Jorabchi, K.; Smith, L.M.; Ramsey, J.M. Integrated microfluidic device for automated single cell analysis using electrophoretic separation and electrospray ionization mass spectrometry. Anal. Chem. 2010, 82, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Li, X.; Mize, T.H.; Sharpe, T.D.; Graziani, E.I.; Abell, C.; Huck, W.T.S. Sensitive, high throughput detection of proteins in individual, surfactant-stabilized picoliter droplets using nanoelectrospray ionization mass spectrometry. Anal. Chem. 2013, 85, 3812–3816. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Wills, Q.F.; Tipping, A.J.; Datta, K.; Mittal, R.; Goldson, A.J.; Sexton, D.W.; Holmes, C.C. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods 2013, 59, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Min, S.K.; Kim, W.Y.; Cho, Y.; Kim, K.S. Fast DNA sequencing with a graphene-based nanochannel device. Nat. Nanotechnol. 2011, 6, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Petriv, O.I.; Kuchenbauer, F.; Delaney, A.D.; Lecault, V.; White, A.; Kent, D.; Marmolejo, L.; Heuser, M.; Berg, T.; Copley, M.; et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc. Natl. Acad. Sci. USA 2010, 107, 15443–15448. [Google Scholar] [CrossRef] [PubMed]
- White, A.K.; VanInsberghe, M.; Petriv, O.I.; Hamidi, M.; Sikorski, D.; Marra, M.A.; Piret, J.; Aparicio, S.; Hansen, C.L. High-throughput microfluidic single-cell RT-qPCR. Proc. Natl. Acad. Sci. USA 2011, 108, 13999–14004. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Freire, V.; Ebert, A.D.; Kalisky, T.; Quake, S.R.; Wu, J.C. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat. Protoc. 2012, 7, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Streets, A.M.; Zhang, X.N.; Cao, C.; Pang, Y.H.; Wu, X.L.; Xiong, L.; Yang, L.; Fu, Y.S.; Zhao, L.; Tang, F.C.; et al. Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 7048–7053. [Google Scholar] [CrossRef] [PubMed]
- Faley, S.L.; Copland, M.; Reboud, J.; Cooper, J.M. Cell chip array for microfluidic proteomics enabling rapid in situ assessment of intracellular protein phosphorylation. Biomicrofluidics 2011, 5, 24106. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.H.; Martin, V.A.; Geahlen, R.L.; Lu, C. One-step extraction of subcellular proteins from eukaryotic cells. Lab Chip 2010, 10, 2046–2048. [Google Scholar] [CrossRef] [PubMed]
- Bockenhauer, S.; Furstenberg, A.; Yao, X.J.; Kobilka, B.K.; Moerner, W.E. Conformational dynamics of single G protein-coupled receptors in solution. J. Phys. Chem. B 2011, 115, 13328–13338. [Google Scholar] [CrossRef] [PubMed]
- Bheda, P.; Schneider, R. Epigenetics reloaded: The single-cell revolution. Trends Cell Biol. 2014, 24, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Hyun, B.R.; McElwee, J.L.; Soloway, P.D. Single molecule and single cell epigenomics. Methods 2015, 72, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulou, E.G.; Elemento, O. Inferring chromatin-bound protein complexes from genome-wide binding assays. Genome Res. 2013, 23, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, A.B.; Gu, H.C.; Bartels, S.J.J.; Zhang, Y.Y.; Matarese, F.; Simmer, F.; Marks, H.; Bock, C.; Gnirke, A.; Meissner, A.; et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012, 22, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Thege, F.I.; Lannin, T.B.; Saha, T.N.; Tsai, S.; Kochman, M.L.; Hollingsworth, M.A.; Rhim, A.D.; Kirby, B.J. Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: Characterization, optimization and downstream analysis. Lab Chip 2014, 14, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Bichsel, C.A.; Gobaa, S.; Kobel, S.; Secondini, C.; Thalmann, G.N.; Cecchini, M.G.; Lutolf, M.P. Diagnostic microchip to assay 3D colony-growth potential of captured circulating tumor cells. Lab Chip 2012, 12, 2313–2316. [Google Scholar] [CrossRef] [PubMed]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.I.; Morgan, B.; Trautwein, J.; et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, 179ra47. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.A.; Talasaz, A.H.; Zhang, H.Y.; Coram, M.A.; Reddy, A.; Deng, G.; Telli, M.L.; Advani, R.H.; Carlson, R.W.; Mollick, J.A.; et al. Single cell profiling of circulating tumor cells: Transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE 2012, 7, e33788. [Google Scholar] [CrossRef] [PubMed]
- Golkowski, M.; Brigham, J.L.; Perera, B.G.K.; Romano, G.S.; Maly, D.J.; Ong, S.E. Rapid profiling of protein kinase inhibitors by quantitative proteomics. Medchemcomm 2014, 5, 363–369. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo, S.-J.; Yao, D.-J. Get to Understand More from Single-Cells: Current Studies of Microfluidic-Based Techniques for Single-Cell Analysis. Int. J. Mol. Sci. 2015, 16, 16763-16777. https://doi.org/10.3390/ijms160816763
Lo S-J, Yao D-J. Get to Understand More from Single-Cells: Current Studies of Microfluidic-Based Techniques for Single-Cell Analysis. International Journal of Molecular Sciences. 2015; 16(8):16763-16777. https://doi.org/10.3390/ijms160816763
Chicago/Turabian StyleLo, Shih-Jie, and Da-Jeng Yao. 2015. "Get to Understand More from Single-Cells: Current Studies of Microfluidic-Based Techniques for Single-Cell Analysis" International Journal of Molecular Sciences 16, no. 8: 16763-16777. https://doi.org/10.3390/ijms160816763






