Inhibition of Mammalian Target of Rapamycin Complex 1 (mTORC1) Downregulates ELOVL1 Gene Expression and Fatty Acid Synthesis in Goat Fetal Fibroblasts
Abstract
:1. Introduction
2. Results
2.1. cDNA Cloning and Sequence Analysis
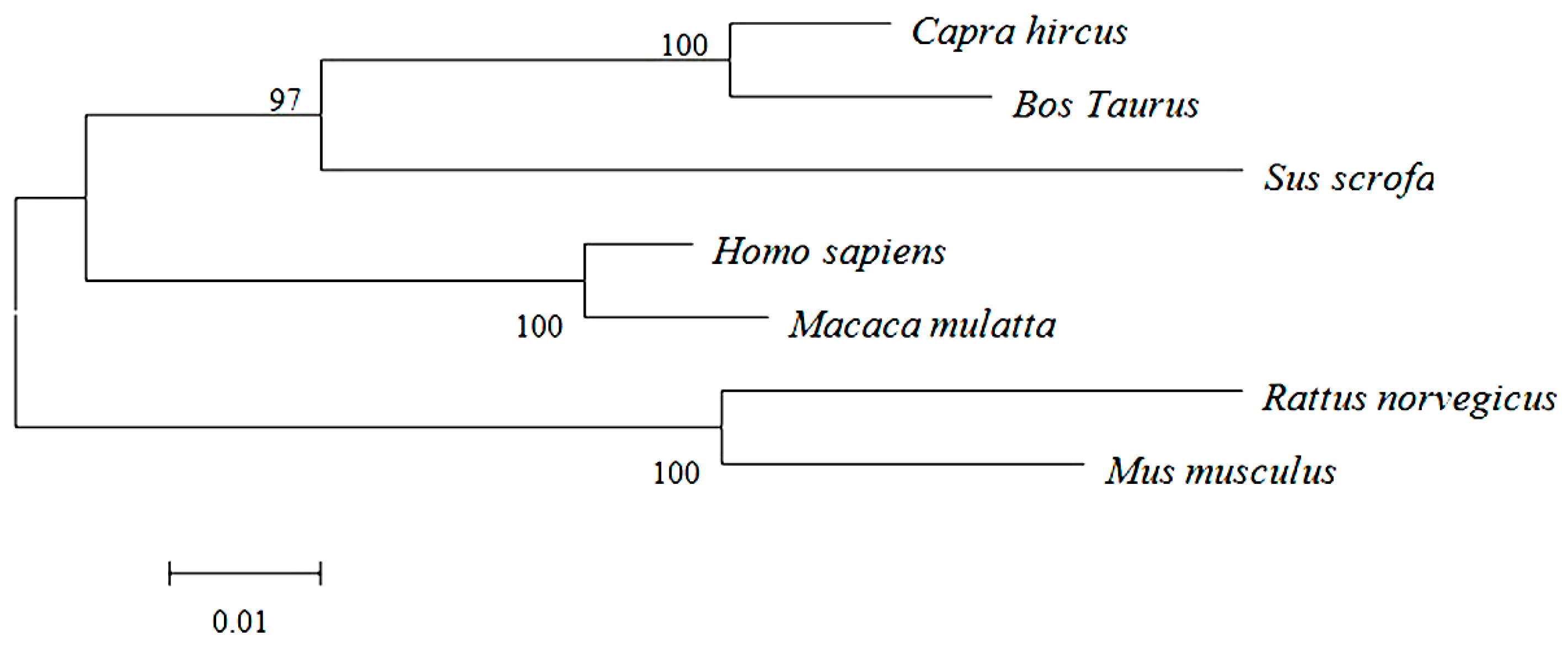
2.2. Primary and Secondary Structure of Cashmere Goat ELOVL1 Protein


2.3. Tissue Distribution of Cashmere Goat ELOVL1 mRNA

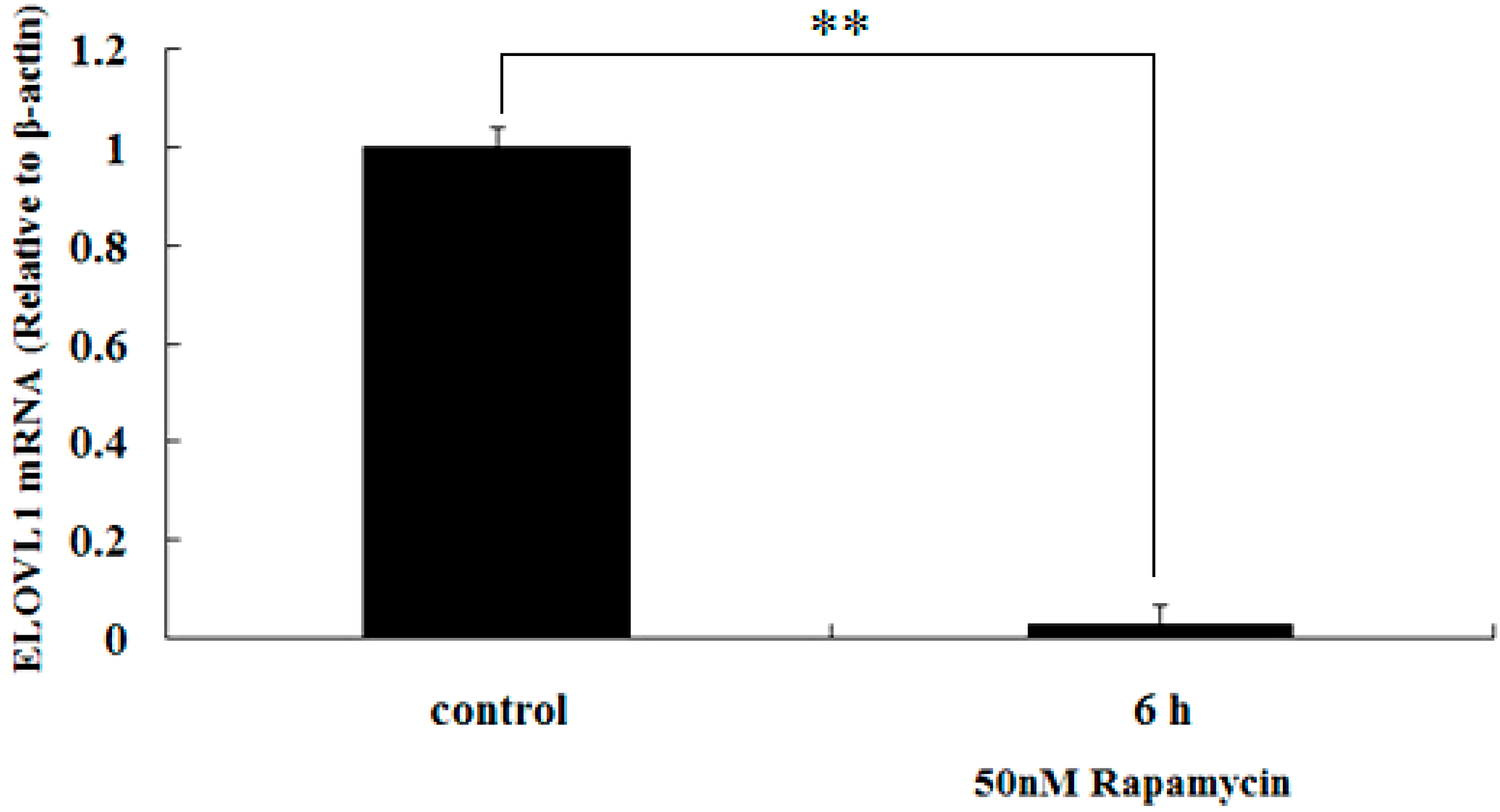
2.4. Rapamycin Down-Regulates the Transcription of ELOVL1 in GFb Cells
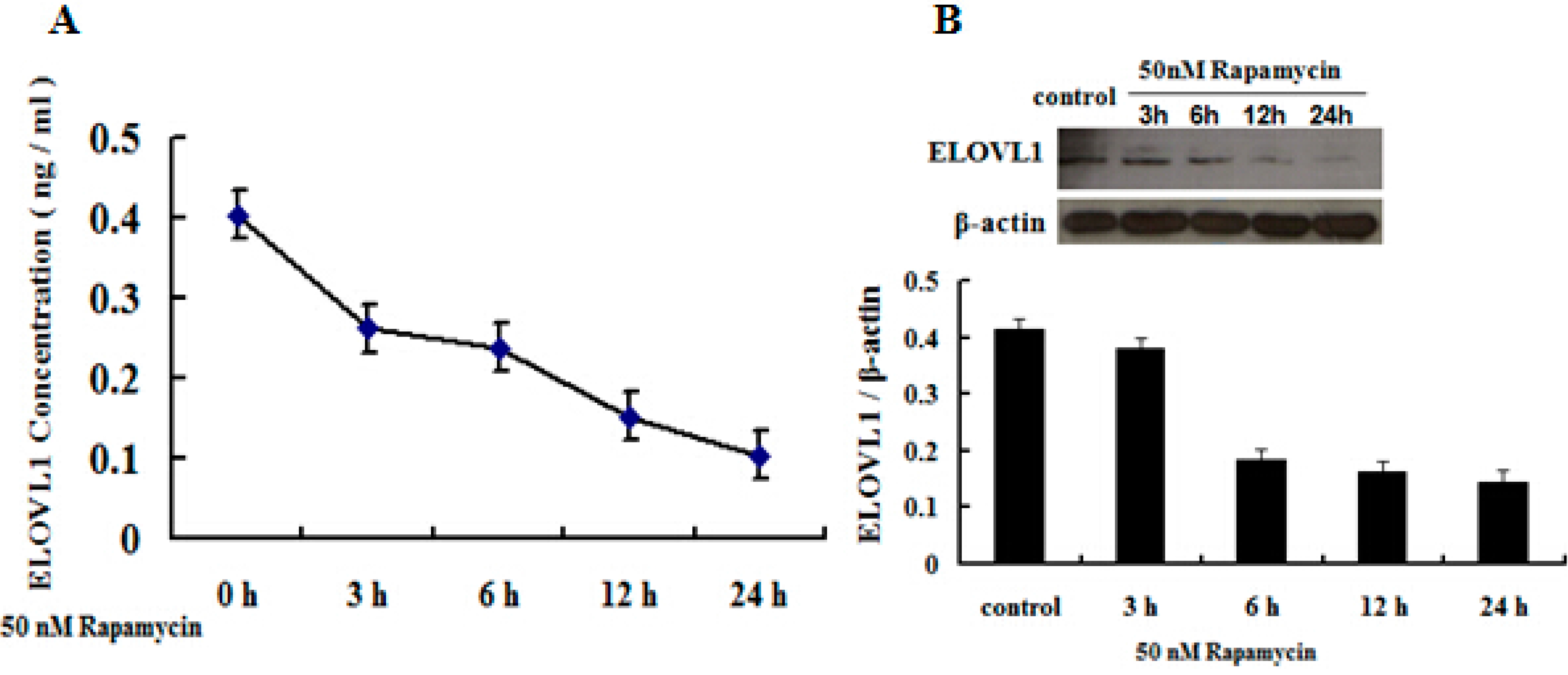
2.5. Rapamycin Attenuates ELOVL1 Expression and Fatty Acid Synthesis in GFb Cells
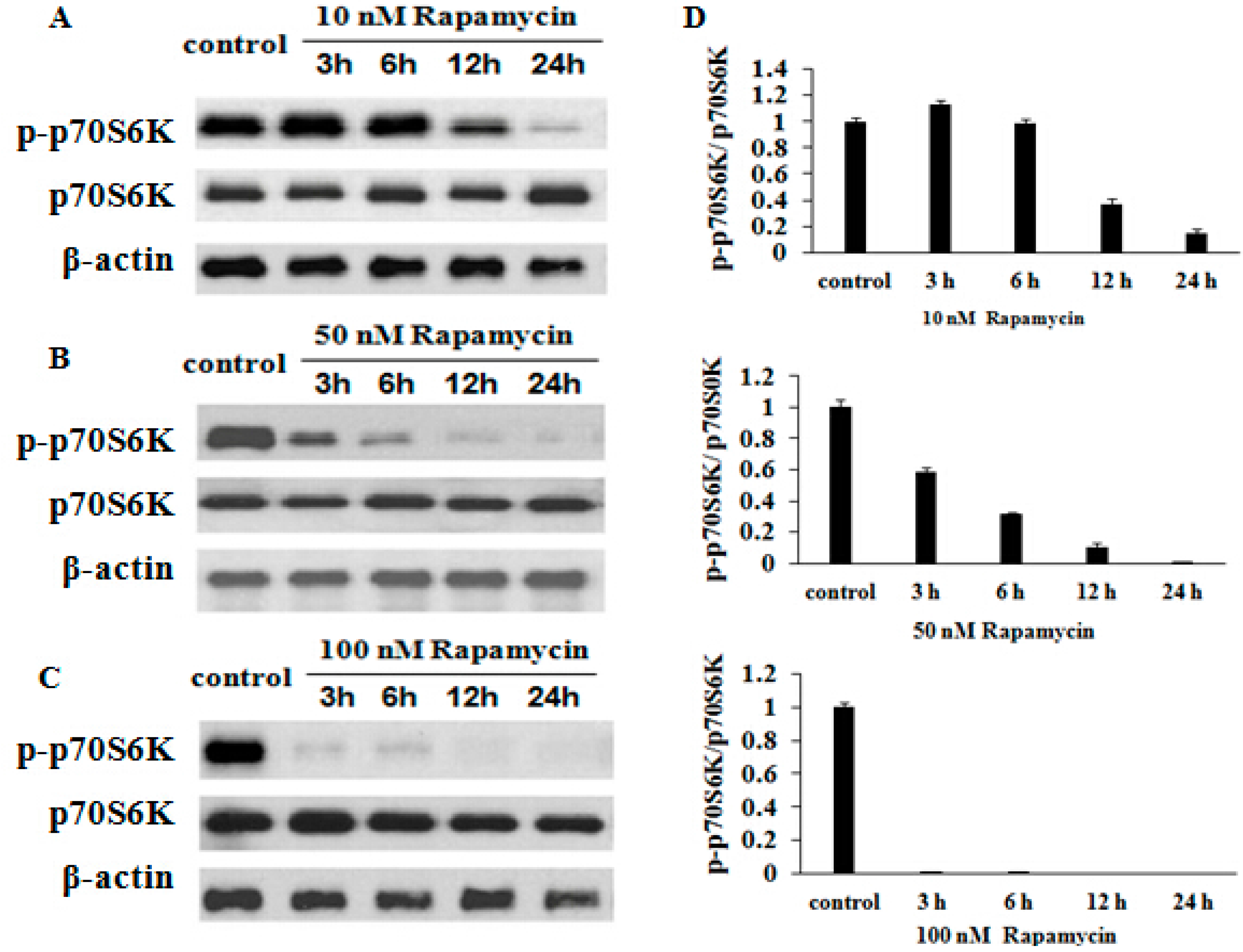
2.6. Rapamycin Inhibits p70S6K (Thr 389) Expression in a Time- and Dose-Dependent Manner
| Fatty Acid | Control (mg/kg) | Treatment (mg/kg) |
|---|---|---|
| Undecanedioic acid (11:0) | 0.12 ± 0.004 | 0.11 ± 0.009 |
| Tridecanoic acid (13:0) | 0.04 ± 0.008 | 0.04 ± 0.008 |
| Myristic acid (14:0) | 4.62 ± 0.48 | 4.21 ± 0.29 |
| Pentadecanoic acid (15:0) | 0.44 ± 0.01 | 0.41 ± 0.03 |
| Palmitic acid (16:0) | 606.42 ± 22.50 | 565.10 ± 32.51 |
| Margaric acid (17:0) | 4.07 ± 0.02 | 3.84 ± 0.35 |
| Stearic acid (18:0) | 721.06 ± 36.82 | 664.16 ± 30.31 |
| Oleic acid (18:1) | 7.16 ± 1.97 | 6.45 ± 0.62 |
| Linoleic acid (18:2) | 1.36 ± 0.24 | 1.37 ± 0.28 |
| Arachidic acid (20:0) | 3.72 ± 0.14 | 3.34 ± 0.27 |
| Heneicosanic acid (21:0) | 0.58 ± 0.04 | 0.55 ± 0.05 |
| Behenic acid (22:0) | 0.22 ± 0.02 | 0.19 ± 0.03 |
| Erucic acid (22:1) | 2.36 ± 0.15 | 2.13 ± 0.25 |
| Tricosanoic acid (23:0) | 0.03 ± 0.01 | 0.01 ± 0.004 |
3. Discussion
4. Experimental Section
4.1. Animal and Tissue Collection
4.2. Cell Culture Conditions
4.3. Total RNA Extraction and Full-Length cDNA Isolation
4.4. Cloning and Sequencing of ELOVL1 cDNA
4.5. Tissue Distribution Analysis and Relative Abundance in GFb Cells of ELOVL1 mRNA by Real-Time q-PCR
4.6. Bioinformatics Analysis
4.7. ELISA
4.8. Gas Chromatography and Mass Spectrum
4.9. Western Blot and Antibodies
4.10. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ELOVL1 | elongation of very-long-chain fatty acids 1 |
| DHA | docosahexaenoic acid |
| EPA | eicosapentaenoic acid |
| ELO | yeast elongase |
| GFb cells | goat fetal fibroblasts |
| mTOR | mammalian target of rapamycin |
| PPAR | peroxisome proliferator-activated receptor |
| and SREBP1 | sterol regulatory element-binding protein 1 |
References
- Kihara, A. Very long-chain fattyacids: Elongation, physiology and realated disorders. J. Biochem. 2012, 152, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Sassa, T.; Kihara, A. Metabolisem of very long-chain Fatty acids: Genes and pathophysiology. Biomol. Ther. 2014, 22, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Suto, S.; Yamanaka, M.; Mizutani, Y.; Mitsutake, S.; Igarashi, Y.; Sassa, T.; Kihara, A. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 18439–18444. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Schackmann, M.J.; Ofman, R.; Dijkstra, I.M.; Wanders, R.J.; Kemp, S. Enzymatic characterization of ELOVL1, a key enzyme in very long-chain fatty acid synthesis. Biochim. Biophys. Acta 2015, 1851, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.E.; Bobik, G.; Dorado, J.; Kroeger, P.E.; Chuang, L.T.; Thurmond, J.M.; Parker-Barnes, J.M.; Das, T.; Huang, Y.S.; Mukerji, P. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 2000, 350, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Tvrdik, P.; Westerberg, R.; Silve, S.; Asadi, A.; Jakobsson, A.; Cannon, B.; Loison, G.; Jacobsson, A. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J. Cell Biol. 2000, 149, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, D.; Wegner, M.S.; Wanger, R.A.; Ferreirós, N.; Schreiber, Y.; Lucks, J.; Schiffmann, S.; Geisslinger, G.; Grösch, S. The equilibrium between long and very long chain ceramides is important for the fate of the cell and can beinfluenced by co-expression of CerS. Int. J. Biochem. Cell Biol. 2013, 45, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Sassa, T.; Wakashima, T.; Ohno, Y.; Kihara, A. Lorenzo’s oil inhibits ELOVL1 and lowers the level of sphingomyelin with a saturated very long-chain fatty acid. J. Lipid Res. 2014, 55, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Düvel, K.; Yecies, J.L.; Menon, S.; Raman, P.; Lipovsky, A.I.; Souza, A.L.; Triantafellow, E.; Cleaver, S. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 2010, 39, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, I.J.; Wei, X.; Semenkovich, C.F. Lipoexpediency: De novo lipogenesis as a metabolic signal transmitter. Trends Endocrinol. 2011, 22, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lamming, D.W.; Sabatini, D.M. A central role for mTOR in lipid homeostasis. Cell Metab. 2013, 18, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Denic, V.; Weissman, J.S. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 2007, 130, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Riezman, H. The long and short of fatty acid synthesis. Cell 2007, 130, 587–588. [Google Scholar] [CrossRef] [PubMed]
- Monroig, Ó.; Guinot, D.; Hontoria, F.; Tocher, D.R.; Navarro, J.C. Biosynthesis of essential fatty acids in Octopus vulgaris (Cuvier, 1797): Molecular cloning, functional characterisation and tissue distribution of a fatty acyl elongase. Aquaculture 2012, 360, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Oh, C.S.; Toke, D.A.; Mandala, S.; Martin, C.E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 1997, 272, 17376–17384. [Google Scholar] [CrossRef] [PubMed]
- Ofman, R.; Dijkstra, I.M.E. The role of ELOVL1 in very long-chain fatty acid homeostasis and X-linked adrenoleukodystrophy. EMBO Mol. Med. 2010, 2, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 2013, 126, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norrmén, C.; Figlia, G.; Lebrun-Julien, F.; Pereira, J.A.; Trötzmüller, M.; Köfeler, H.C.; Rantanen, V.; Wessig, C.; van Deijk, A.L.; Smit, A.B.; et al. mTORC1 controls PNS myelination along the mTORC1-RXRγ-SREBP-lipid biosynthesis axis in Schwann cells. Cell Rep. 2014, 9, 646–660. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lin, Y.; Bian, Y.; Liu, L.; Shao, L.; Lin, L.; Qu, B.; Zhao, F.; Gao, X.; Li, Q. Leucyl-tRNA synthetase regulates lactation and cell proliferation via mTOR signaling in dairy cow mammary epithelial cells. Int. J. Mol. Sci. 2014, 15, 5952–5969. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tikhonenko, M.; Bozack, S.N.; Lydic, T.A.; Yan, L.; Panchy, N.L.; McSorley, K.M.; Faber, M.S.; Yan, Y.; Boulton, M.E.; et al. Changes in the daily rhythm of lipid metabolism in the diabetic retina. PLoS ONE 2014, 9, e95028. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Antoñanzas, G.; Tocher, D.R.; Martinez-Rubio, L.; Leaver, M.J. Conservation of lipid metabolic gene transcriptional regulatory networks in fish and mammals. Gene 2014, 534, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Botolin, D.; Xu, J.; Christian, B.; Mitchell, E.; Jayaprakasam, B.; Nair, M.G.; Peters, J.M.; Busik, J.V.; Olson, L.K.; et al. Regulation of hepatic fatty acid elongase and desaturase expression in diabetes and obesity. J. Lipid Res. 2006, 47, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Kuo, S.H.; Hsu, C.H.; Chen, L.T.; Lu, Y.S; Lin, C.H.; Yeh, P.Y.; Jeng, H.J.; Gao, M.; Yeh, K.H.; Cheng, A.L. Lack of compensatory pAKT activation and eIF4E phosphorylation of lymphoma cells towards mTOR inhibitor, RAD001. Eur. J. Cancer 2011, 17, 1244–1257. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Shan, C.; Liu, L.; Zhou, T.; Zhou, J.; Hu, X.; Chen, Y.; Cui, H.; Gao, N. Tanshinone IIA inhibits HIF-1α and VEGF expression in breast cancer cells via mTOR/p70S6K/RPS6/4E-BP1 signaling pathway. PLoS ONE 2015, 10, e0117440. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, W.; Zheng, M.; Xue, Y.; Yang, J.; Liu, W.; Han, S. The mTOR inhibitor rapamycin synergizes with a fatty acid synthase inhibitor to induce cytotoxicity in ER/HER2-positive breast cancer cells. PLoS ONE 2014, 27, e97697. [Google Scholar]
- Basic Local Alignment Search Tool. Available online: http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 4 June 2013).
- Compute pI/Mw tool. Available online: http://web.expasy.org/compute_pi/ (accessed on 4 June 2013).
- PSORT WWW Server. Available online: http://psort.hgc.jp/ (accessed on 7 July 2013).
- SMART. Available online: http://smart.embl-heidelberg.de/ (accessed on 28 October 2013).
- The Pfam database. Available online: http://pfam.xfam.org/ (accessed on 28 October 2013).
- SoftBerry-Psite. Available online: http://www.softberry.com/ (accessed on 7 July 2013).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; He, Q.; Guo, Z.; Yang, L.; Bao, L.; Bao, W.; Zheng, X.; Wang, Y.; Wang, Z. Inhibition of Mammalian Target of Rapamycin Complex 1 (mTORC1) Downregulates ELOVL1 Gene Expression and Fatty Acid Synthesis in Goat Fetal Fibroblasts. Int. J. Mol. Sci. 2015, 16, 16440-16453. https://doi.org/10.3390/ijms160716440
Wang W, He Q, Guo Z, Yang L, Bao L, Bao W, Zheng X, Wang Y, Wang Z. Inhibition of Mammalian Target of Rapamycin Complex 1 (mTORC1) Downregulates ELOVL1 Gene Expression and Fatty Acid Synthesis in Goat Fetal Fibroblasts. International Journal of Molecular Sciences. 2015; 16(7):16440-16453. https://doi.org/10.3390/ijms160716440
Chicago/Turabian StyleWang, Weipeng, Qiburi He, Zhixin Guo, Limin Yang, Lili Bao, Wenlei Bao, Xu Zheng, Yanfeng Wang, and Zhigang Wang. 2015. "Inhibition of Mammalian Target of Rapamycin Complex 1 (mTORC1) Downregulates ELOVL1 Gene Expression and Fatty Acid Synthesis in Goat Fetal Fibroblasts" International Journal of Molecular Sciences 16, no. 7: 16440-16453. https://doi.org/10.3390/ijms160716440





