Stress-Responsive Expression, Subcellular Localization and Protein–Protein Interactions of the Rice Metacaspase Family
Abstract
:1. Introduction
2. Results
2.1. Expression Patterns of OsMCs in Response to Abiotic Stresses
2.1.1. Expression Patterns in Response to Drought and Salt Stresses
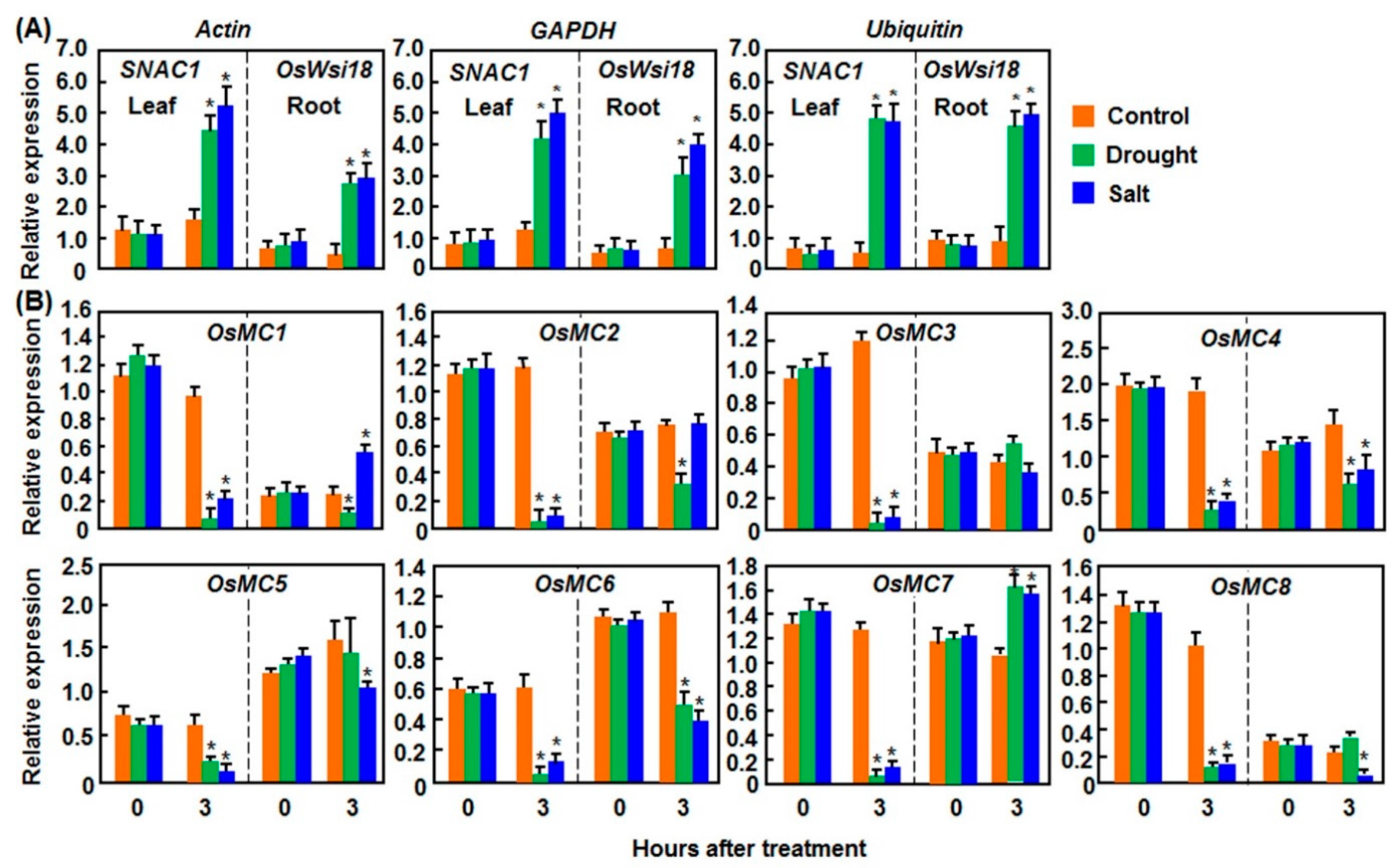
2.1.2. Expression Patterns in Response to Cold and Heat Stresses
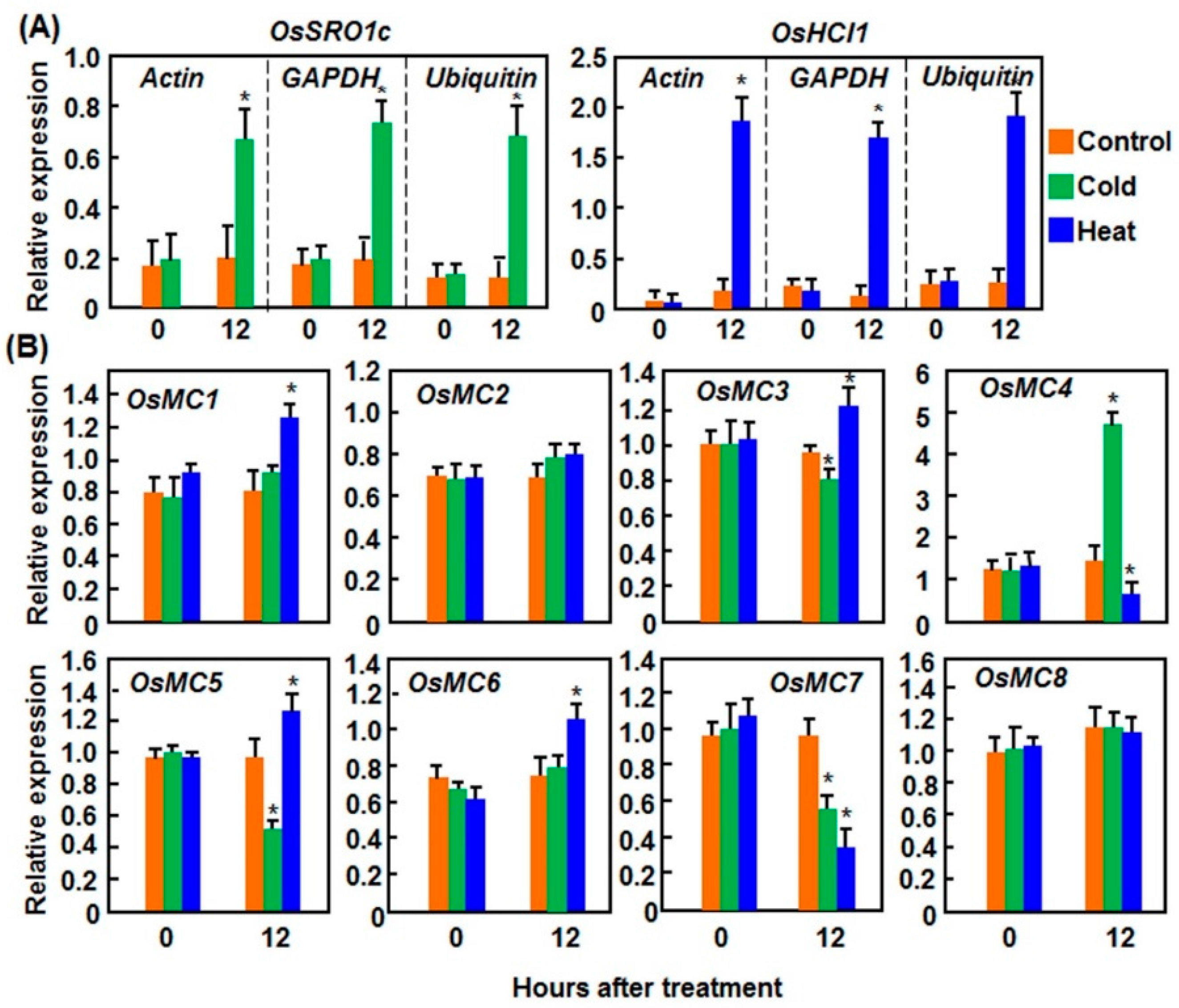
2.2. Expression Patterns of OsMCs in Response to Pathogens
2.2.1. Expression Patterns in Response to M. oryzae

2.2.2. Expression Patterns in Response to Xoo
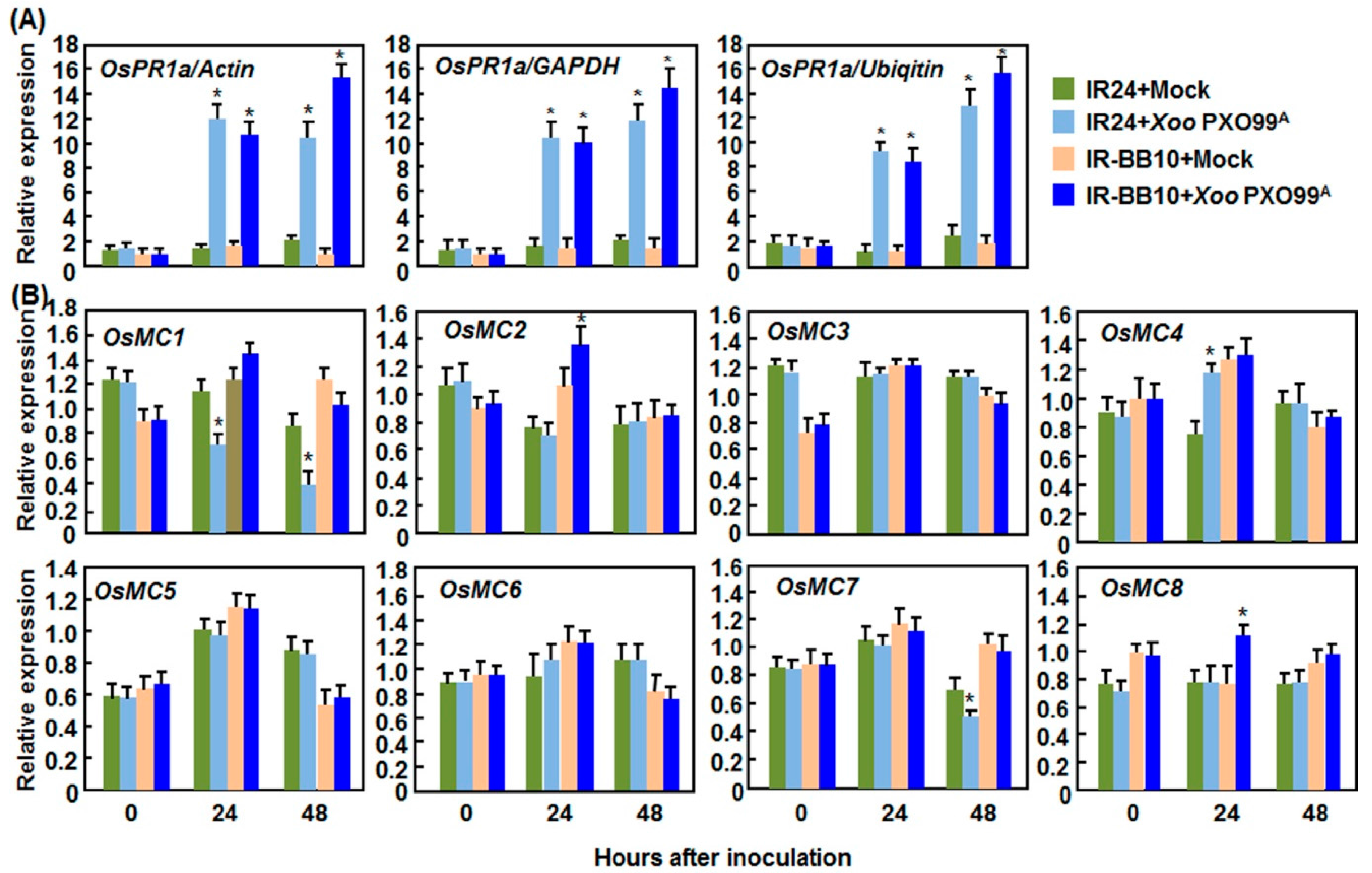
2.2.3. Expression Patterns in Response to R. solani
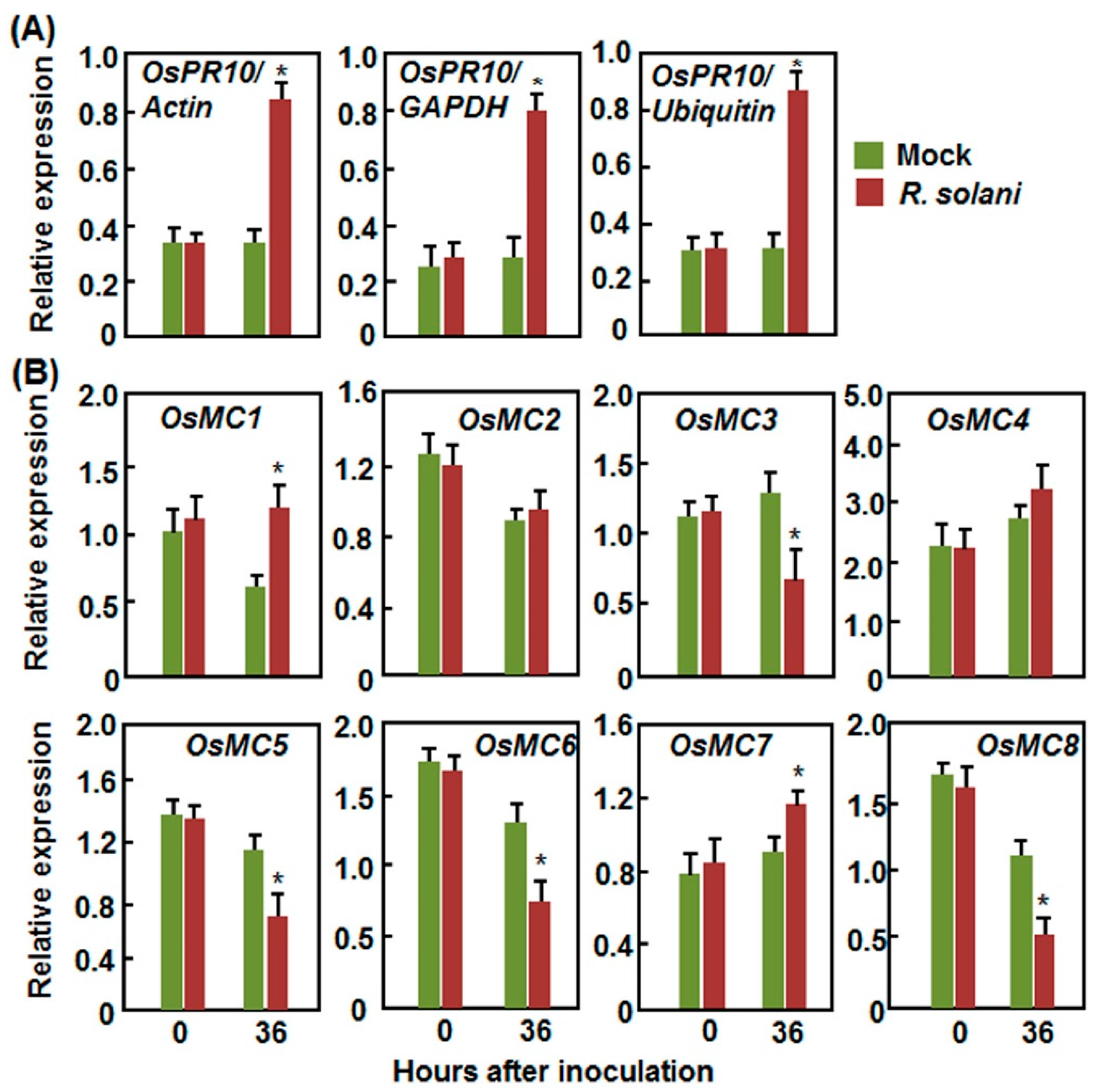
2.3. Expression Patterns of OsMCs in Response to Stress-Related Hormones
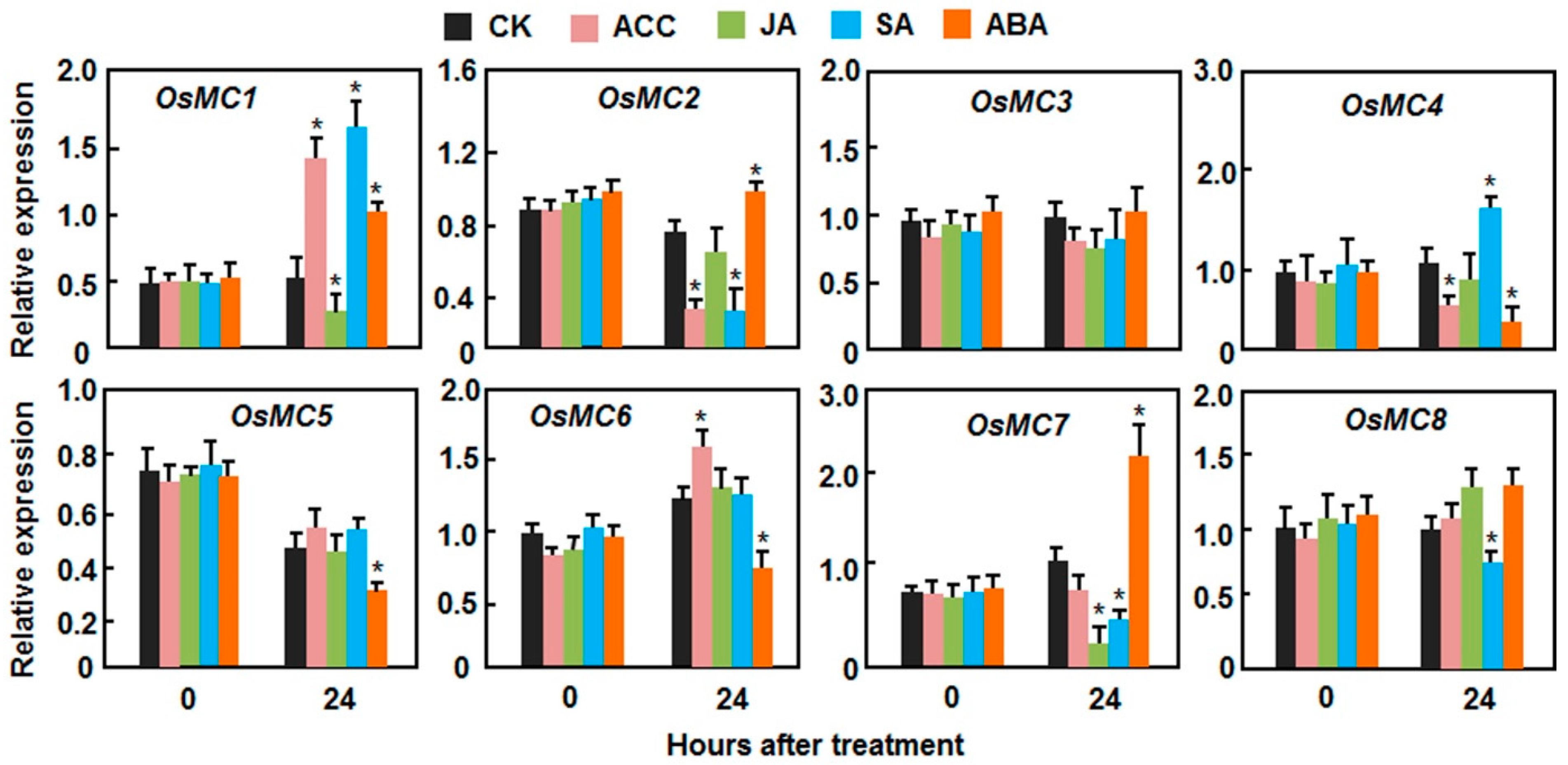
2.4. Putative Cis-Elements in Promoter Regions of the OsMC Genes
| Regulator | Cis-Element | Code | Numbers of cis-Elements | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OsMC1 | OsMC2 | OsMC3 | OsMC4 | OsMC5 | OsMC6 | OsMC7 | OsMC8 | |||
| ABA | ABRELATERD1 | S000414 | – | 2 | 1 | – | – | 2 | – | 1 |
| ABREOSRAB21 | S000012 | – | – | 2 | – | – | – | – | – | |
| ABRERATCAL | S000408 | – | 3 | – | – | – | 2 | – | 1 | |
| MYB | MYBCORE | S000176 | 2 | 4 | 5 | 1 | 1 | 2 | 1 | 1 |
| MYC | MYCCONSENSUSAT | S000407 | 4 | 14 | 8 | 4 | 4 | 8 | 6 | 8 |
| WRKY | WBOXATNPR1 | S000390 | 1 | 1 | 1 | – | 4 | – | – | 3 |
| WBOXNTERF3 | S000457 | 2 | 1 | 1 | 1 | 2 | – | 1 | 6 | |
| WRKY710S | S000447 | 3 | 4 | 3 | 4 | 6 | 1 | 1 | 10 | |
| ERF | GCCCORE | S000430 | – | – | 3 | 1 | 2 | 2 | – | 1 |
| Auxin | NTBBF1ARROLB | S000273 | 1 | – | – | – | 1 | – | – | – |
2.5. Subcellular Localization of OsMCs
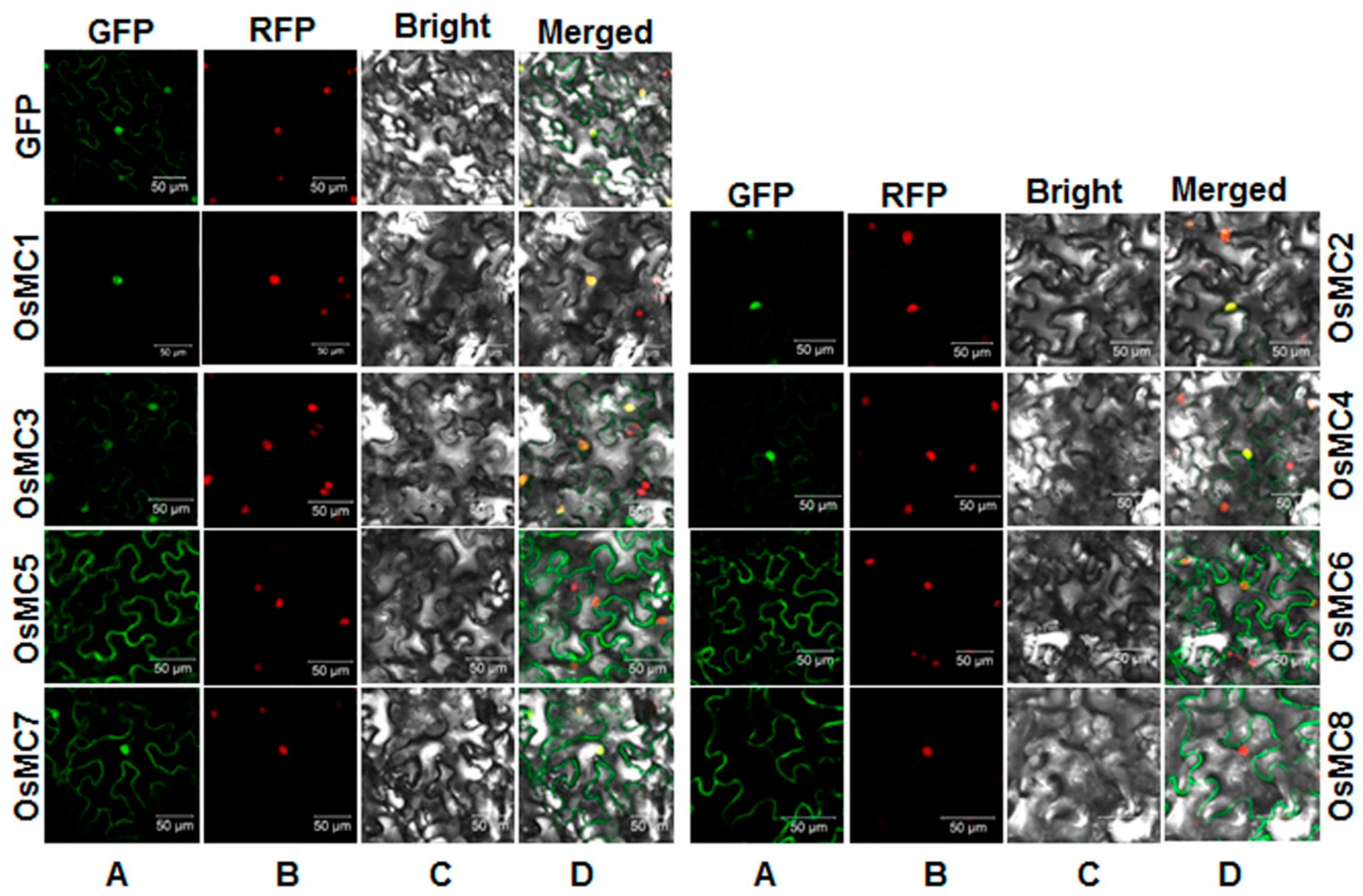
2.6. Interaction Relationships between Type I OsMCs and OsLSDs
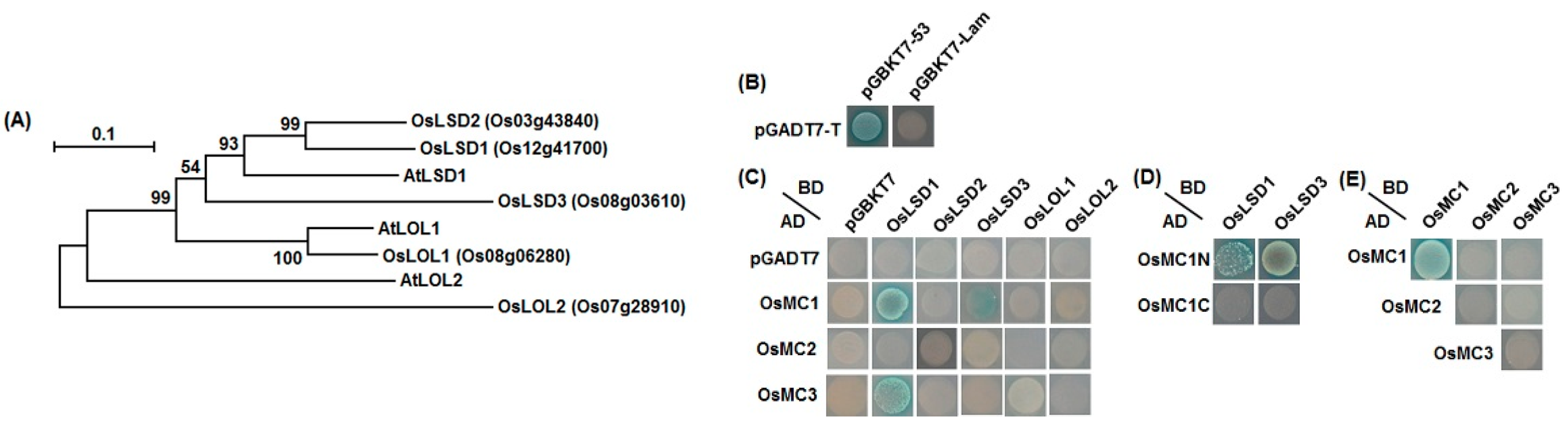
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth
4.2. Treatments with Abiotic Stress and Hormones
4.3. Inoculation with Different Pathogens
4.4. qRT-PCR Analysis of Gene Expression
| Primers | Sequences (5′-3′) | Size (bp) |
|---|---|---|
| Cloning of cDNAs | ||
| OsMC1-F | ATGGATCACTTCGGCGGACG | 1107 |
| OsMC1-R | TTACAGGACGAACGGCTTGC | |
| OsMC2-F | ATGGCGAGCGCGAGGCCGCC | 1110 |
| OsMC2-R | TCACAAGAGGAAGGGCTTCC | |
| OsMC3-F | ATGGGCTGCAACTGCCTCGT | 1203 |
| OsMC3-R | TCACAGGAGAAACGGTTTCC | |
| OsMC4-F | ATGGGGCGGAAGAGAGCGGT | 1230 |
| OsMC4-R | TTAGCATATGAAAGCCACGT | |
| OsMC5-F | ATGGGGGGCCGGAAGCGCGC | 1263 |
| OsMC5-R | TCAGCATATGAAGGCCACAC | |
| OsMC6-F | ATGGGCCGCAAGCGCGCGCT | 1254 |
| OsMC6-R | TCAGCATATAAAAGACACAT | |
| OsMC7-F | ATGGAGAGGGGTCAGAAGAA | 1026 |
| OsMC7-R | TCAGAGCGCCGTCATGGCCT | |
| OsMC8-F | ATGGCGGTCGTCAGCGGCGG | 909 |
| OsMC8-R | TCACAGGATAAACTGCTCCT | |
| OsLSD1-F | ATGTGCATTGCTGAACCAAT | 606 |
| OsLSD1-R | TCAGCTGCTGGGCTTCTGGT | |
| OsLSD2-F | ATGGTGGCTTCAAGAGCTCCA | 444 |
| OsLSD2-R | CTATCCTAGACTGAAAAGCA | |
| OsLSD3-F | ATGCAGAGCCAGATCGTGT | 519 |
| OsLSD3-R | TTACTTTTTACCACCAGTTGTA | |
| OsLSD4-F | ATGCAGAGCCAGATCGTGTG | 561 |
| OsLSD4-R | CTATTTCCCAGTTGTAACTCCA | |
| OsLSD5-F | ATGCAGGACCAGCTGATCTG | 444 |
| OsLSD5-R | TCATCTTTTCCATGAGGTGAC | |
| Subcellular Localization Assays | ||
| OsMC1-GFP-F | CGCGGATCC ATGGATCACTTCGGCGGACG | 1107 |
| OsMC1-GFP-R | TGCTCTAGA TTACAGGACGAACGGCTTGC | |
| OsMC2-GFP-F | CGCGGATCC ATGGCGAGCGCGAGGCCGCC | 1110 |
| OsMC2-GFP-R | TGCTCTAGA TCACAAGAGGAAGGGCTTCC | |
| OsMC3-GFP-F | CGCGGATCC ATGGGCTGCAACTGCCTCGT | 1203 |
| OsMC3-GFP-R | TGCTCTAGA TCACAGGAGAAACGGTTTCC | |
| OsMC4-GFP-F | CGCGGATCC ATGGGGCGGAAGAGAGCGGT | 1230 |
| OsMC4-GFP-R | TGCTCTAGA TTAGCATATGAAAGCCACGT | |
| OsMC5-GFP-F | CGCGGATCC ATGGGGGGCCGGAAGCGCGC | 1263 |
| OsMC5-GFP-R | TGCTCTAGA TCAGCATATGAAGGCCACAC | |
| OsMC6-GFP-F | CGCGGATCC ATGGGCCGCAAGCGCGCGCT | 1254 |
| OsMC6-GFP-R | TGCTCTAGA TCAGCATATAAAAGACACAT | |
| OsMC7-GFP-F | CGCGGATCC ATGGAGAGGGGTCAGAAGAA | 1026 |
| OsMC7-GFP-R | TGCTCTAGA TCAGAGCGCCGTCATGGCCT | |
| OsMC8-GFP-F | CGCGGATCC ATGGCGGTCGTCAGCGGCGG | 909 |
| OsMC8-GFP-R | TCCCCCGGG TCACAGGATAAACTGCTCCT | |
| qRT-PCR Assays | ||
| OsMC1-RT-F | GCTTCATCAAGGCGGTGGAGTG | 142 |
| OsMC1-RT-R | AAGTTGGCGACCTTGCGGATG | |
| OsMC2-RT-F | CGACCCGTACAGGGTGCCGA | 166 |
| OsMC2-RT-R | GCACAGCGCCTCGTCGTAGC | |
| OsMC3-RT-F | GGCTCCTTCGTCCGCAAGAT | 101 |
| OsMC3-RT-R | CACAGGAGAAACGGTTTCCTGT | |
| OsMC4-RT-F | TCGACGTTCGTGGAGATGCTC | 126 |
| OsMC4-RT-R | ATTCACGAGCCGCCTGATCTT | |
| OsMC5-RT-F | GTGCCAGACCGACCAGACAT | 102 |
| OsMC5-RT-R | CCGCTCTTCTCCGACAGGAT | |
| OsMC1-RT-F | GCTTCATCAAGGCGGTGGAGTG | 142 |
| OsMC1-RT-R | AAGTTGGCGACCTTGCGGATG | |
| OsMC6-RT-F | CCACACCGCAGGGTTCTTCAT | 147 |
| OsMC6-RT-R | GTCCAGGCTGCTGAGTGTATCC | |
| OsMC7-RT-F | ATACAGACCGTGCTGGCGTC | 143 |
| OsMC7-RT-R | AGGAATGGCGTCTCGGCGTT | |
| OsMC8-RT-F | TCCGGCAAGTGCCTCGTAAC | 150 |
| OsMC8-RT-R | CAATGCGGTCGGTCACAGGAT | |
| Yeast Two-Hybrid Assays | ||
| OsMC1-BD-F | CGGAATTC ATGGATCACTTCGGCGGACGT | 1107 |
| OsMC1-BD-R | CGCGGATCC TTACAGGACGAACGGCTTGCG | |
| OsMC2-BD-F | CCGGAATTC ATGGCGAGCGCGAGGCCG | 1110 |
| OsMC2-BD-R | CGCGGATCC TCACAAGAGGAAGGGCTTC | |
| OsMC3-BD-F | CCGGAATTC ATGGGCTGCAACTGCCTCGTC | 1203 |
| OsMC3-BD-R | CGCGGATCC TCACAGGAGAAACGGTTTCC | |
| OsMC1-BDN-R | TGCTCTAGA CTTGCCGCGGGAGCCCGGGA | 252 |
| OsMC1-BDC-F | CGCGGATCC AAGCGCGCCGTCCTGATCGGC | 855 |
| OsLSD1-AD-F | CCGGAATTC ATGTGCATTGCTGAACCAAT | 606 |
| OsLSD1-AD-R | CGCGGATCC TCAGCTGCTGGGCTTCTGGT | |
| OsLSD2-AD-F | CCGGAATTC ATGGTGGCTTCAAGAGCTCCA | 444 |
| OsLSD2-AD-R | CGCGGATCC CTATCCTAGACTGAAAAGCA | |
| OsLSD3-AD-F | CCGGAATTC ATGCAGAGCCAGATCGTGT | 519 |
| OsLSD3-AD-R | CGCGGATCC TTACTTTTTACCACCAGTTGTA | |
| OsLSD4-AD-F | CCGGAATTC ATGCAGAGCCAGATCGTGTG | 561 |
| OsLSD4-AD-R | CGCGGATCC CTATTTCCCAGTTGTAACTCCA | |
| OsLSD5-AD-F | CCGGAATTC ATGCAGGACCAGCTGATCTG | 444 |
| OsLSD5-AD-R | CGCGGATCC TCATCTTTTCCATGAGGTGAC | |
4.5. Subcellular Localization of the OsMC Proteins
4.6. Yeast Two-Hybrid Interaction Assays
4.7. Bioinformatics Analysis of Cis-Element in the Promoters of the OsMC Genes
4.8. Accession Numbers
5. Conclusions
| Genes | M. oryzae | Xoo | R. solani | Abiotic Stress | Hormones | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compatible | Incompatible | Compatible | Incompatible | Drought | Salt | Heat | Cold | ACC | JA | SA | ABA | ||
| OsMC1 | ↓ | ↑ | ↓ | -- | ↑ | ↓ | ↑ | ↑ | -- | ↑ | ↓ | ↑ | ↑ |
| OsMC2 | -- | -- | -- | ↑ | -- | ↓ | -- | -- | -- | ↓ | -- | ↓ | ↑ |
| OsMC3 | ↓ | ↑ | -- | -- | ↓ | -- | -- | ↑ | ↓ | -- | -- | -- | -- |
| OsMC4 | ↓ | -- | ↑ | -- | -- | ↓ | ↓ | ↓ | ↑ | ↓ | -- | ↑ | ↓ |
| OsMC5 | ↑ | ↑ | -- | -- | ↓ | -- | ↓ | ↑ | ↓ | -- | -- | -- | ↓ |
| OsMC6 | -- | ↑ | -- | -- | ↓ | ↓ | ↓ | ↑ | -- | ↑ | -- | -- | ↓ |
| OsMC7 | ↑ | ↑ | ↓ | -- | ↑ | ↑ | ↑ | ↓ | ↓ | -- | ↓ | ↓ | ↑ |
| OsMC8 | ↓ | -- | -- | ↑ | ↓ | -- | ↓ | -- | -- | -- | -- | ↓ | -- |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kroemer, G.; El-Deiry, W.S.; Golstein, P.; Peter, M.E.; Vaux, D.; Vandenabeele, P.; Zhivotovsky, B.; Blagosklonny, M.V.; Malorni, W.; Knight, R.A.; et al. Classification of cell death: Recommendations of the nomenclature committee on cell death. Cell Death Differ. 2005, 12, 1463–1467. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.M. Caspases: The executioners of apoptosis. Biochem. J. 1997, 326, 1–16. [Google Scholar] [PubMed]
- Kitanaka, C.; Kuchino, Y. Caspase-independent programmed cell death with necrotic morphology. Cell Death Differ. 1999, 6, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Martin, S.J. Caspase-independent cell death. Nat. Med. 2005, 11, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. Classification of the caspase-hemoglobinase fold: Detection of new families and implications for the origin of the eukaryotic separins. Proteins 2002, 46, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Martins, L.M.; Kaufmann, S.H. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999, 68, 383–424. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.G.; O’Rourke, K.; Aravind, L.; Pisabarro, M.T.; Seshagiri, S.; Koonin, E.V.; Dixit, V.M. Identification of paracaspases and metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 2000, 6, 961–967. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P.V. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xue, Q. Molecular phylogeny, evolution, and functional divergence of the LSD1-like gene family: Inference from the rice genome. J. Mol. Evol. 2007, 64, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.K.; Zhang, H. Genome wide survey and characterization of metacaspase gene family in rice (Oryza sativa). J. Genet. 2014, 93, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Gong, P.; Wei, R.; Li, S.; Zhang, X.; Yu, Y.; Wang, Y. The metacaspase gene family of Vitis vinifera L.: Characterization and differential expression during ovule abortion in stenospermocarpic seedless grapes. Gene 2013, 528, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Maspons, A.; Sepulveda-Garcia, E.; Sanchez-Baldoquin, L.; Marrero-Gutierrez, J.; Pons, T.; Rocha-Sosa, M.; Gonzalez, L. Two aspartate residues at the putative p10 subunit of a type II metacaspase from Nicotiana tabacum L. may contribute to the substrate-binding pocket. Planta 2014, 239, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Tsiatsiani, L.; Timmerman, E.; de Bock, P.J.; Vercammen, D.; Stael, S.; van de Cotte, B.; Staes, A.; Goethals, M.; Beunens, T.; van Damme, P.; et al. The Arabidopsis metacaspase9 degradome. Plant Cell 2013, 25, 2831–2847. [Google Scholar] [CrossRef] [PubMed]
- Wrzaczek, M.; Vainonen, J.P.; Stael, S.; Tsiatsiani, L.; Help-Rinta-Rahko, H.; Gauthier, A.; Kaufholdt, D.; Bollhoner, B.; Lamminmaki, A.; Gevaert, K.; et al. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 2015, 34, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lam, E. Sheathing the swords of death: Post-translational modulation of plant metacaspases. Plant Signal. Behav. 2011, 6, 2051–2056. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.; Zhang, Y. Regulating the reapers: Activating metacaspases for programmed cell death. Trends Plant Sci. 2012, 17, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Vercammen, D.; van de Cotte, B.; de Jaeger, G.; Eeckhout, D.; Casteels, P.; Vandepoele, K.; Vandenberghe, I.; van Beeumen, J.; Inze, D.; van Breusegem, F. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 2004, 279, 45329–45336. [Google Scholar] [CrossRef] [PubMed]
- Vercammen, D.; Belenghi, B.; van de Cotte, B.; Beunens, T.; Gavigan, J.A.; de Rycke, R.; Brackenier, A.; Inze, D.; Harris, J.L.; van Breusegem, F. Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. J. Mol. Biol. 2006, 364, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Belenghi, B.; Romero-Puertas, M.C.; Vercammen, D.; Brackenier, A.; Inze, D.; Delledonne, M.; van Breusegem, F. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J. Biol. Chem. 2007, 282, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Ma, Q.M.; Zhang, Y.L.; Yang, J.P.; Zhao, G.H.; Fu, D.Q.; Luo, Y.B.; Qu, G.Q. Biochemical evidence of key residues for the activation and autoprocessing of tomato type II metacaspase. FEBS Lett. 2013, 587, 2517–2522. [Google Scholar] [CrossRef] [PubMed]
- Bozhkov, P.V.; Suarez, M.F.; Filonova, L.H.; Daniel, G.; Zamyatnin, A.A., Jr.; Rodriguez-Nieto, S.; Zhivotovsky, B.; Smertenko, A. Cysteine protease mc II-Pa executes programmed cell death during plant embryogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 14463–14468. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem. 2005, 280, 14691–14699. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 2011, 66, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Piszczek, E.; Dudkiewicz, M.; Mielecki, M. Biochemical and bioinformatic characterization of type II metacaspase protein (TaeMCA II) from wheat. Plant Mol. Biol. Rep. 2012, 30, 1338–1347. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Drury, G.E.; Rotari, V.I.; Gordon, A.; Willer, M.; Farzaneh, T.; Woltering, E.J.; Gallois, P. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J. Biol. Chem. 2008, 283, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Hoeberichts, F.A.; ten Have, A.; Woltering, E.J. A tomato metacaspase gene is upregulated during programmed cell death in Botrytis cinerea-infected leaves. Planta 2003, 217, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Bae, C.; Oh, S.K.; Choi, D. A pepper (Capsicum annuum L.) metacaspase 9 (Camc9) plays a role in pathogen-induced cell death in plants. Mol. Plant Pathol. 2013, 14, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Wang, X.J.; Feng, H.; Tang, C.L.; Bai, P.F.; Wei, G.R.; Huang, L.L.; Kang, Z.S. TaMCA4, a novel wheat metacaspase gene functions in programmed cell death induced by the fungal pathogen Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 2012, 25, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Goodwin, P.H.; Hsiang, T. Expression of a metacaspase gene of Nicotiana benthamiana after inoculation with Colletotrichum destructivum or Pseudomonas syringae pv. tomato, and the effect of silencing the gene on the host response. Plant Cell Rep. 2007, 26, 1879–1888. [Google Scholar] [PubMed]
- Ahmad, R.; Zuily-Fodil, Y.; Passaquet, C.; Bethenod, O.; Roche, R.; Repellin, A. Ozone and aging up-regulate type II metacaspase gene expression and global metacaspase activity in the leaves of field-grown maize (Zea. mays L.) plants. Chemosphere 2012, 87, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. Recent advance in the study of caspase-like proteases and Bax inhibitor-1 in plants: Their possible roles as regulator of programmed cell death. Mol. Plant Pathol. 2004, 5, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Bollhoner, B.; Zhang, B.; Stael, S.; Denance, N.; Overmyer, K.; Goffner, D.; van Breusegem, F.; Tuominen, H. Post mortem function of Atmc9 in xylem vessel elements. New Phytol. 2013, 200, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Suarez, M.F.; Filonova, L.H.; Smertenko, A.; Savenkov, E.I.; Clapham, D.H.; von Arnold, S.; Zhivotovsky, B.; Bozhkov, P.V. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr. Biol. 2004, 14, R339–R340. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Filonova, L.H.; Fukada, K.; Savenkov, E.I.; Gogvadze, V.; Clapham, D.; Sanchez-Vera, V.; Suarez, M.F.; Zhivotovsky, B.; Daniel, G.; et al. Autophagy and metacaspase determine the mode of cell death in plants. J. Cell. Biol. 2013, 203, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Smidler, A.; Puigvert, M.; Popa, C.; Valls, M.; Dangl, J.L. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 2014, 21, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, R.A.; Richberg, M.H.; Schmidt, R.; Dean, C.; Dangl, J.L. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 1997, 88, 685–694. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Keunen, E.; Bex, G.J.; Smeets, K.; Vangronsveld, J.; Cuypers, A. Reliable gene expression analysis by reverse transcription-quantitative PCR: Reporting and minimizing the uncertainty in data accuracy. Plant Cell 2014, 26, 3829–3837. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.H.; Dai, M.Q.; Yao, J.L.; Xiao, B.Z.; Li, X.H.; Zhang, Q.F.; Xiong, L.Z. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed]
- Yi, N.; Oh, S.J.; Kim, Y.S.; Jang, H.J.; Park, S.H.; Jeong, J.S.; Song, S.I.; Do Choi, Y.; Kim, J.K. Analysis of the Wsi18, a stress-inducible promoter that is active in the whole grain of transgenic rice. Transgenic Res. 2011, 20, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Zong, W.; Du, H.; Hu, H.H.; Xiong, L.Z. A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses though interaction with various transcription factors. Plant Mol. Biol. 2014, 84, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.D.; Cho, H.Y.; Park, Y.C.; Ham, D.J.; Lee, J.K.; Jang, C.S. The rice RING finger E3 ligase, OsHCI1, drives nuclear export of multiple substrate proteins and its heterogeneous overexpression enhances acquired thermotolerance. J. Exp. Bot. 2013, 64, 2899–2914. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.J.; Zhang, H.J.; Li, D.Y.; Huang, L.; Hong, Y.B.; Ding, X.S.; Nelson, R.S.; Zhou, X.P.; Song, F.M. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 2013, 81, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Rakwal, R.; Jwa, N.S.; Agrawal, V.P. Signalling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: A model illustrating components participating during defence/stress response. Plant Physiol. Biochem. 2001, 39, 1095–1103. [Google Scholar] [CrossRef]
- Young, S.A.; Guo, A.; Guikema, J.A.; White, F.F.; Leach, J.E. Rice cationic peroxidase accumulates in xylem vessels during incompatible interactions with Xanthomonas oryzae pv. oryzae. Plant Physiol. 1995, 107, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhara, I.; Iwai, T.; Seo, S.; Yanagawa, Y.; Kawahigasi, H.; Hirose, S.; Ohkawa, Y.; Ohashi, Y. Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics 2008, 279, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.J.; Wang, A.R.; Shi, Y.J.; Wang, L.Q.; Liu, W.D.; Wang, Z.H.; Lu, G.D. Identification of defense-related genes in rice responding to challenge by Rhizoctonia solani. Theor. Appl. Genet. 2008, 116, 501–516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Jia, W.S.; Yang, J.C.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crop. Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Halim, V.A.; Vess, A.; Scheel, D.; Rosahl, S. The role of salicylic acid and jasmonic acid in pathogen defence. Plant Biol. 2006, 8, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Gontia-Misha, I.; Sasidharan, S.; Tiwari, S. Recent developments in use of 1-aminocyclopropane-1-carboxylate (ACC) deaminase for conferring tolerance to biotic and abiotic stress. Biotechnol. Lett. 2014, 36, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKYsuperfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Chakravarthy, S.; Tuori, R.P.; D’Ascenzo, M.D.; Fobert, P.R.; Despres, C.; Martin, G.B. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 2003, 15, 3033–3050. [Google Scholar] [CrossRef] [PubMed]
- Baumann, K.; de Paolis, A.; Costantino, P.; Gualberti, G. The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the roIB oncogene in plants. Plant Cell 1999, 11, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, R.; Banerjee, R.; Chung, S.M.; Farman, M.; Citovsky, V.; Hogenhout, S.A.; Tzfira, T.; Goodin, M. pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: Probing Nicotiana. benthamiana-virus interactions. Mol. Plant Microbe Interact. 2007, 20, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Epple, P.; Mack, A.A.; Morris, V.R.F.; Dangl, J.L. Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 6831–6836. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Tian, Y.C.; He, C.Z. Cloning of a novel rice gene OsLSD1 and bioinformatic analysis of LSD1-like gene family from Arabidopsis and rice. Prog. Biochem. Biophys. 2005, 32, 268–274. [Google Scholar]
- Wang, L.J.; Pei, Z.Y.; Tian, Y.C.; He, C.Z. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol. Plant Microbe Interact. 2005, 18, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.X.; He, C.Z. The rice OsLOL2 gene encodes a zinc finger protein involved in rice growth and disease resistance. Mol. Genet. Genomics 2007, 278, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.H.; Zhu, C.F.; Pang, J.H.; Zhang, X.R.; Yang, C.L.; Xia, G.X.; Tian, Y.C.; He, C.Z. OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination though the modulation of gibberellin biosynthesis in Oryza sativa. Plant J. 2014, 80, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Oono, Y.; Kanamori, H.; Matsumoto, T.; Itoh, T.; Minami, E. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS ONE 2012, 7, e49423. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.L.; Liu, H.B.; Yuan, B.; Li, X.H.; Xu, C.G.; Wang, S.P. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 2011, 34, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 2009, 379, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Piszczek, E.; Dudkiewicz, M.; Sobczak, M. Molecular cloning and phylogenetic analysis of cereal type II metacaspase cDNA from wheat. Biol. Plant. 2011, 55, 614–624. [Google Scholar] [CrossRef]
- He, S.P.; Huang, K.W.; Zhang, X.; Yu, X.C.; Huang, P.; An, C.C. The LSD1-type zinc finger motifs of Pisum sativa LSD1 are a novel nuclear localization signal and interact with importin α. PLoS ONE 2011, 6, e22131. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Wang, S.P.; Zhang, Q.F. New gene for bacterial blight resistance in rice located on chomosome 12 identified from Minghui 63, an elite restorer line. Phytopathology 2002, 92, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, G.; Park, D.S.; Yang, Y. Inoculation and scoring methods for rice sheath blight disease. Methods Mol. Biol. 2013, 956, 257–268. [Google Scholar] [PubMed]
- Bustin, S.; Beaulieu, J.-F.; Huggett, J.; Jaggi, R.; Kibenge, F.; Olsvik, P.; Penning, L.; Toegel, S. MIQE precis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol. Biol. 2010, 11, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Zhang, H.; Hong, Y.; Liu, S.; Li, D.; Song, F. Stress-Responsive Expression, Subcellular Localization and Protein–Protein Interactions of the Rice Metacaspase Family. Int. J. Mol. Sci. 2015, 16, 16216-16241. https://doi.org/10.3390/ijms160716216
Huang L, Zhang H, Hong Y, Liu S, Li D, Song F. Stress-Responsive Expression, Subcellular Localization and Protein–Protein Interactions of the Rice Metacaspase Family. International Journal of Molecular Sciences. 2015; 16(7):16216-16241. https://doi.org/10.3390/ijms160716216
Chicago/Turabian StyleHuang, Lei, Huijuan Zhang, Yongbo Hong, Shixia Liu, Dayong Li, and Fengming Song. 2015. "Stress-Responsive Expression, Subcellular Localization and Protein–Protein Interactions of the Rice Metacaspase Family" International Journal of Molecular Sciences 16, no. 7: 16216-16241. https://doi.org/10.3390/ijms160716216






