Identifying Similar Patterns of Structural Flexibility in Proteins by Disorder Prediction and Dynamic Programming
Abstract
:1. Introduction
2. Results
2.1. Building up the Dataset
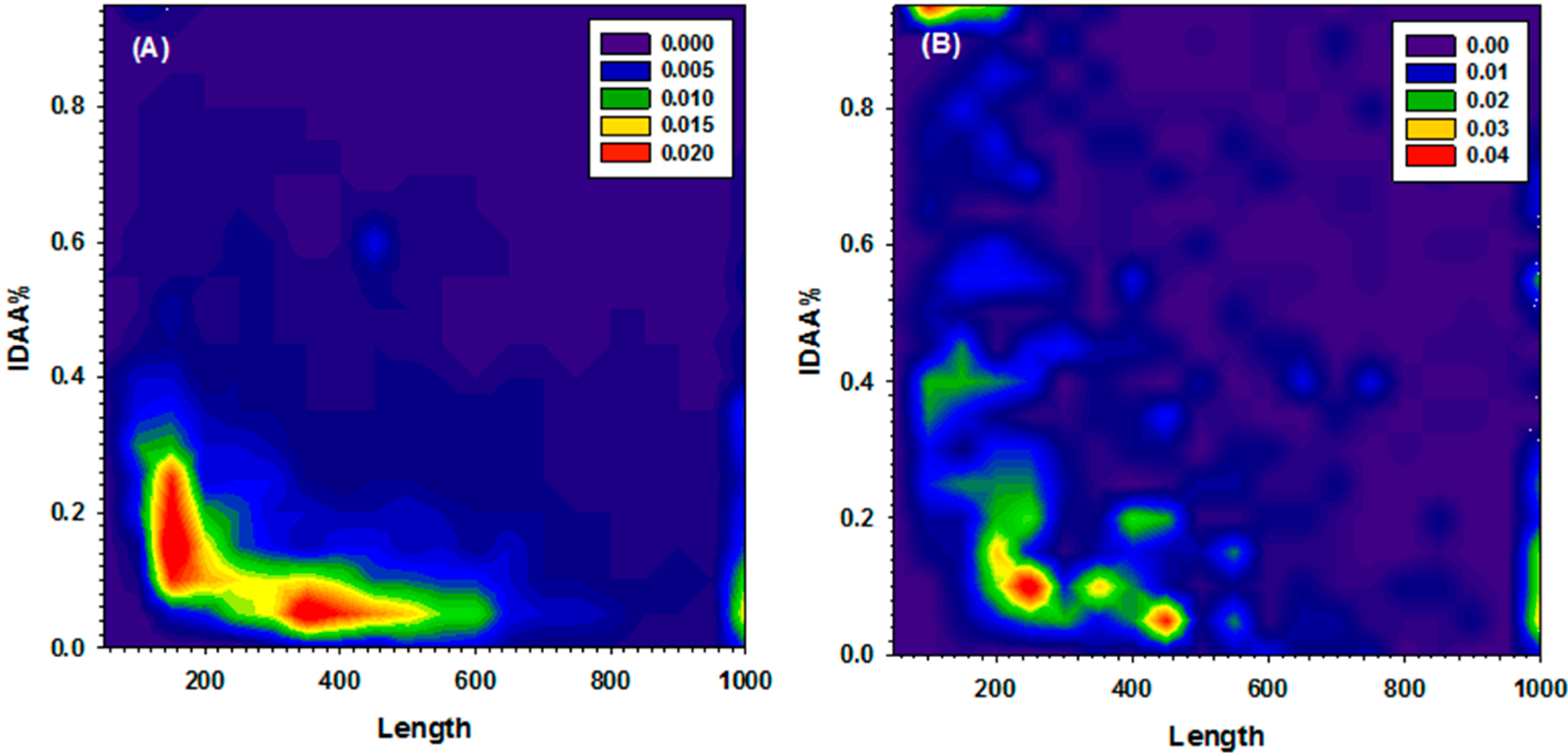
2.2. Gap Penalty

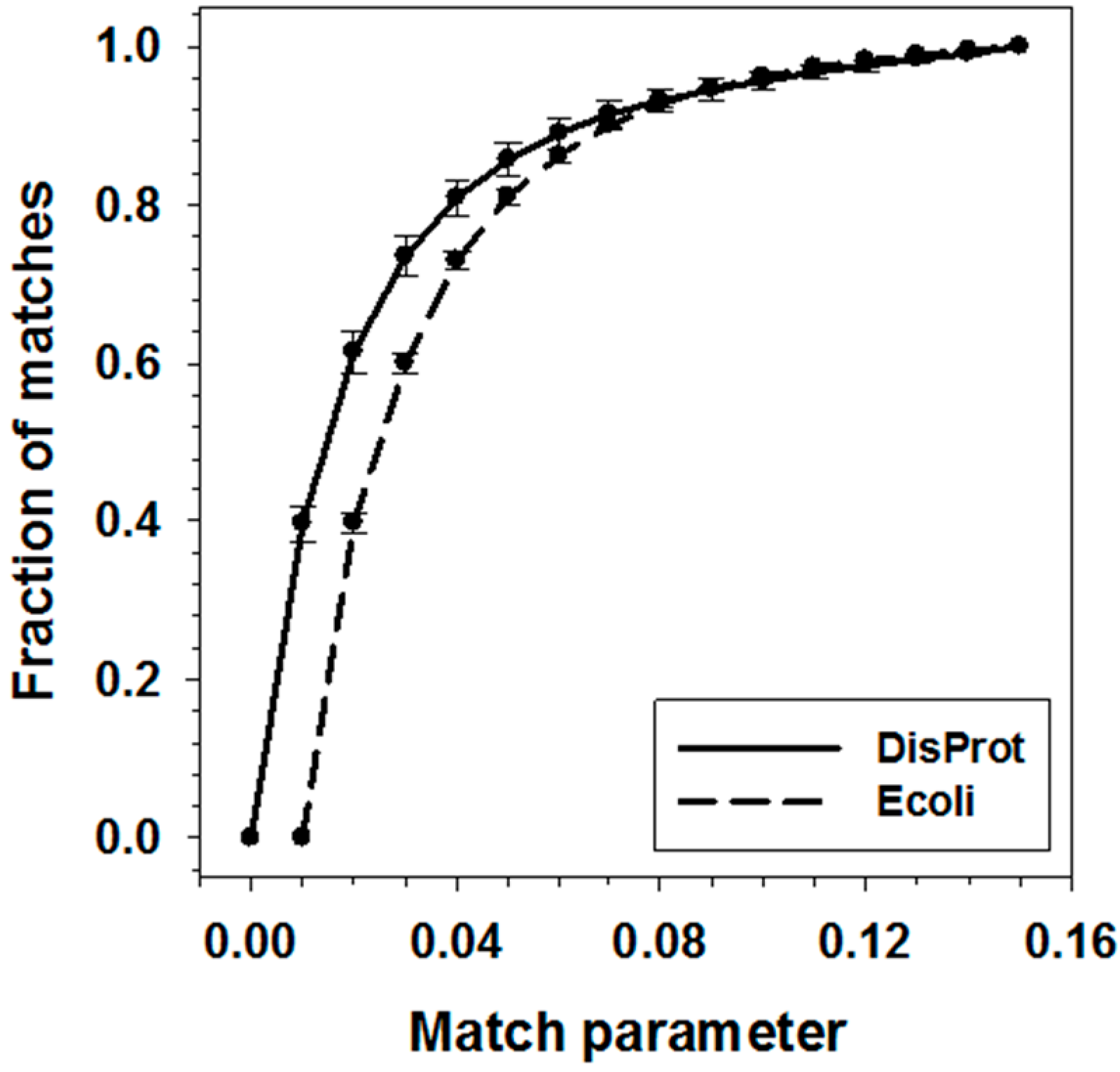
2.3. Match Threshold
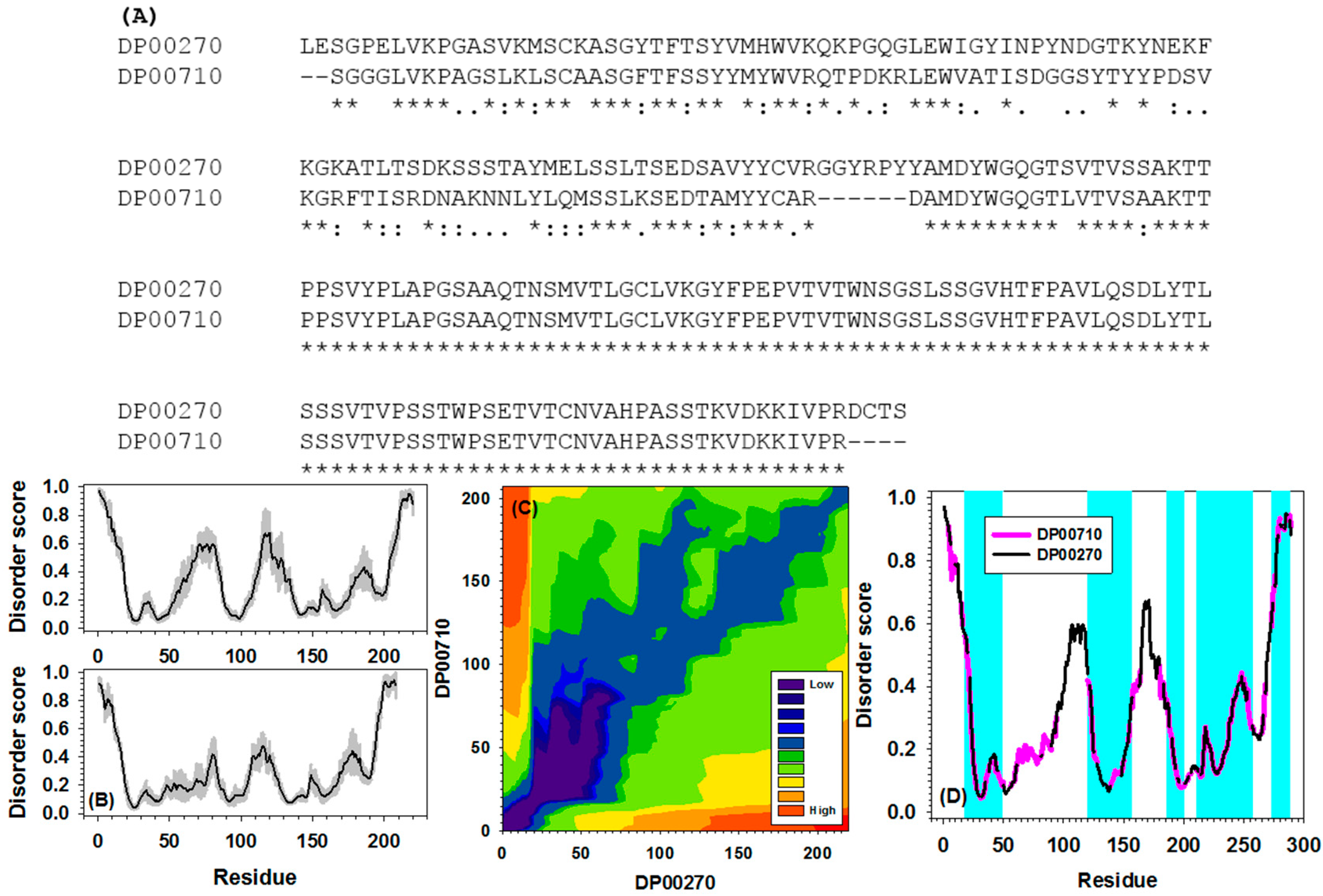
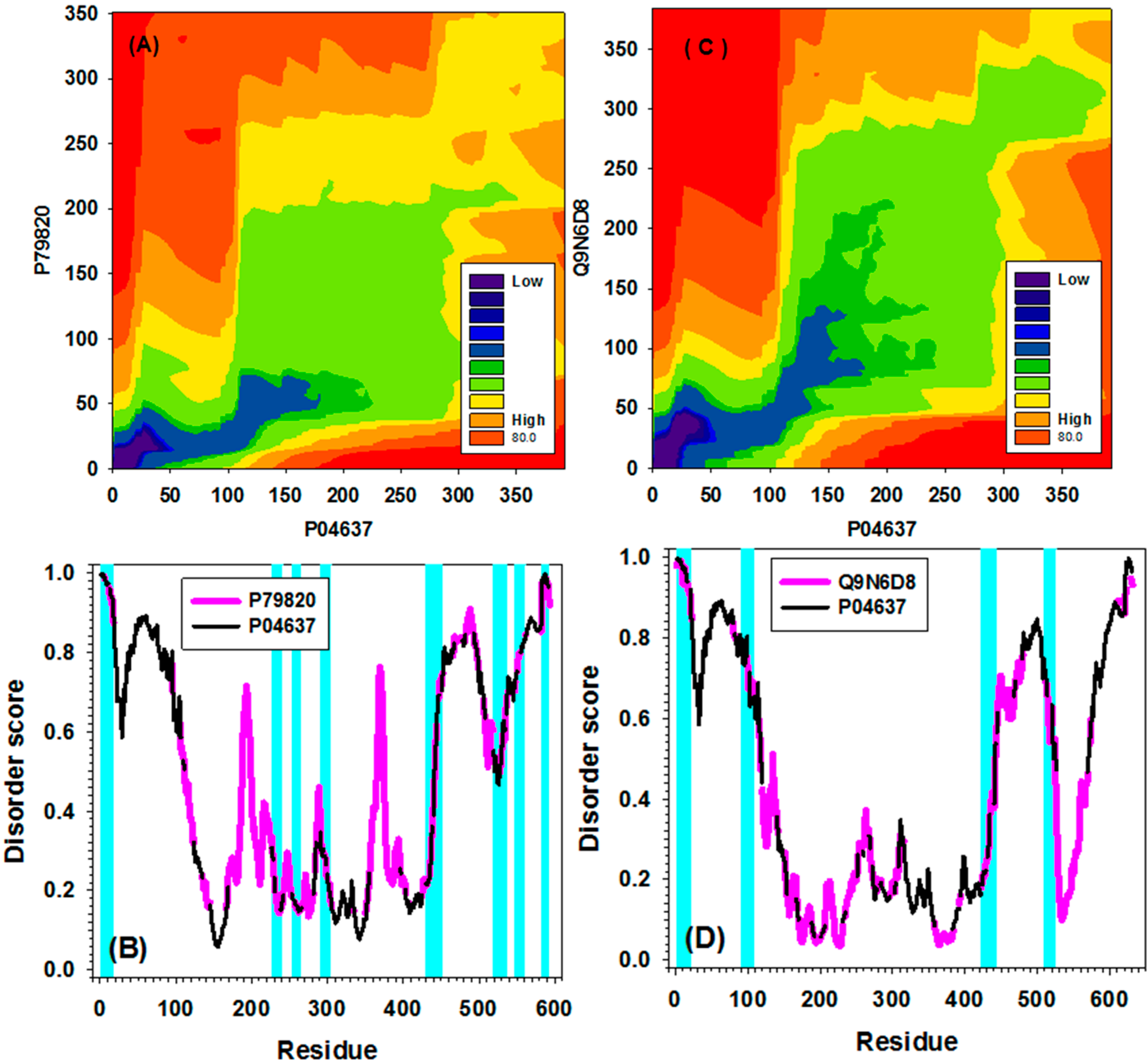
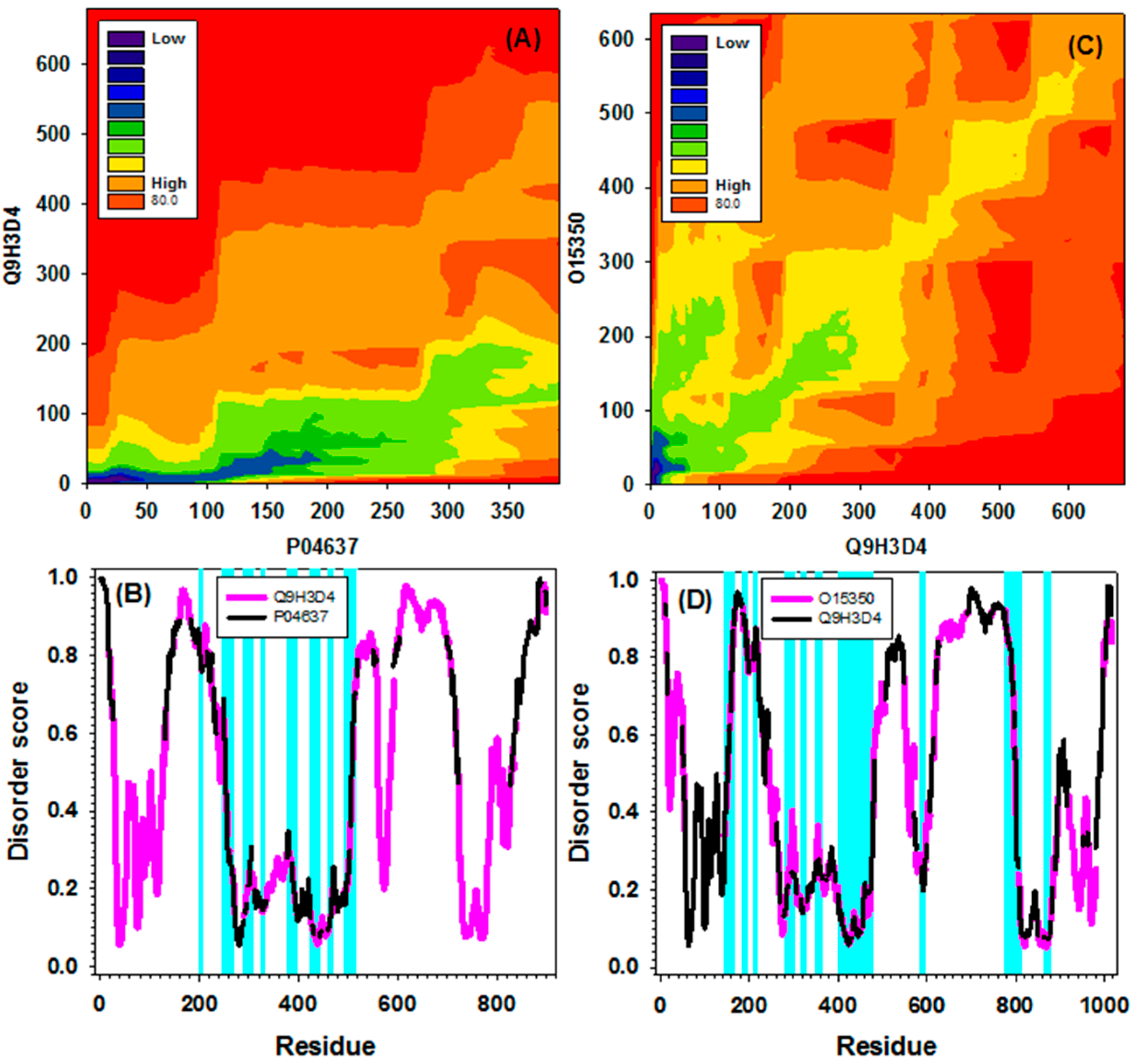
2.4. Examples/Applications

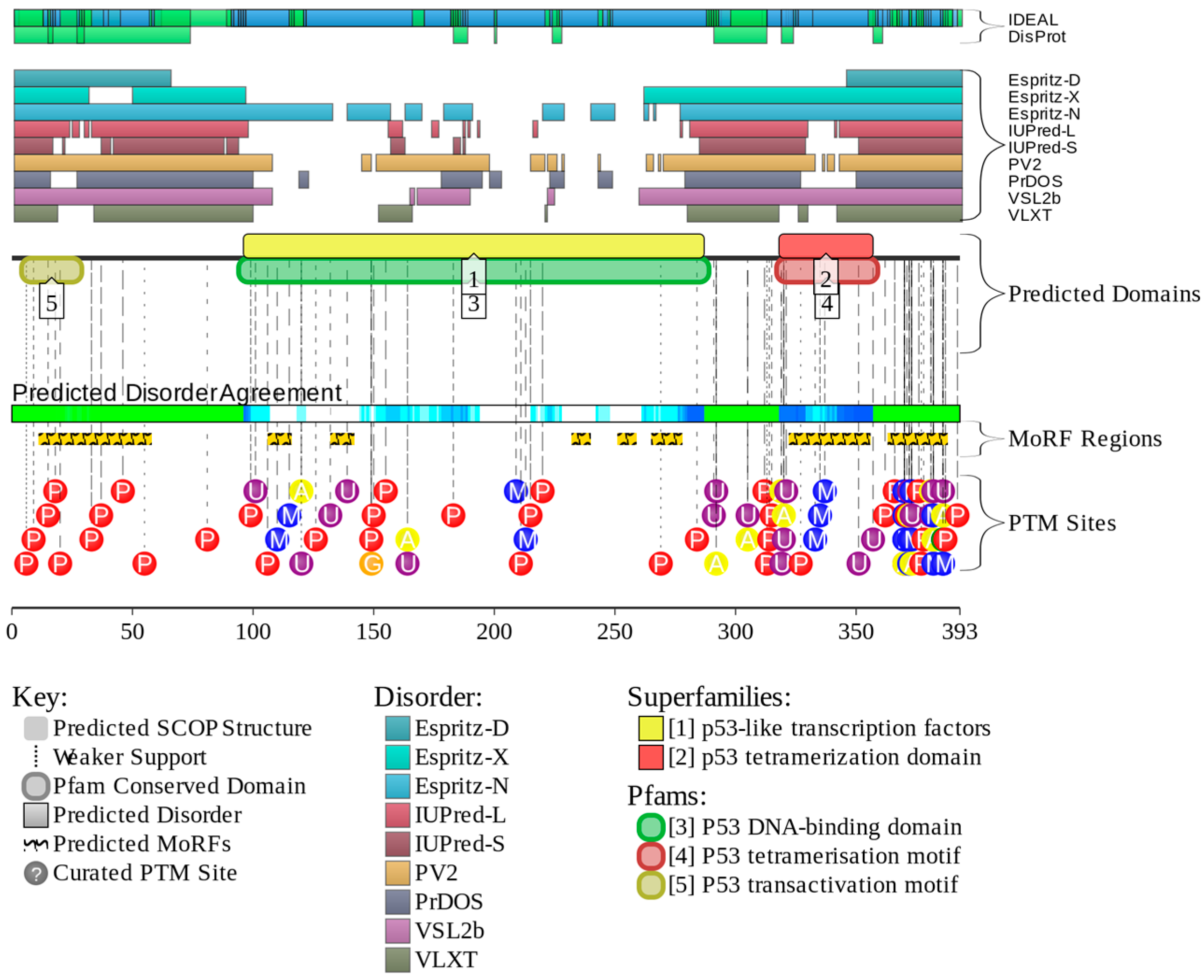
2.5. Web Server

3. Discussions
4. Experimental Section
4.1. Datasets and Disorder Prediction
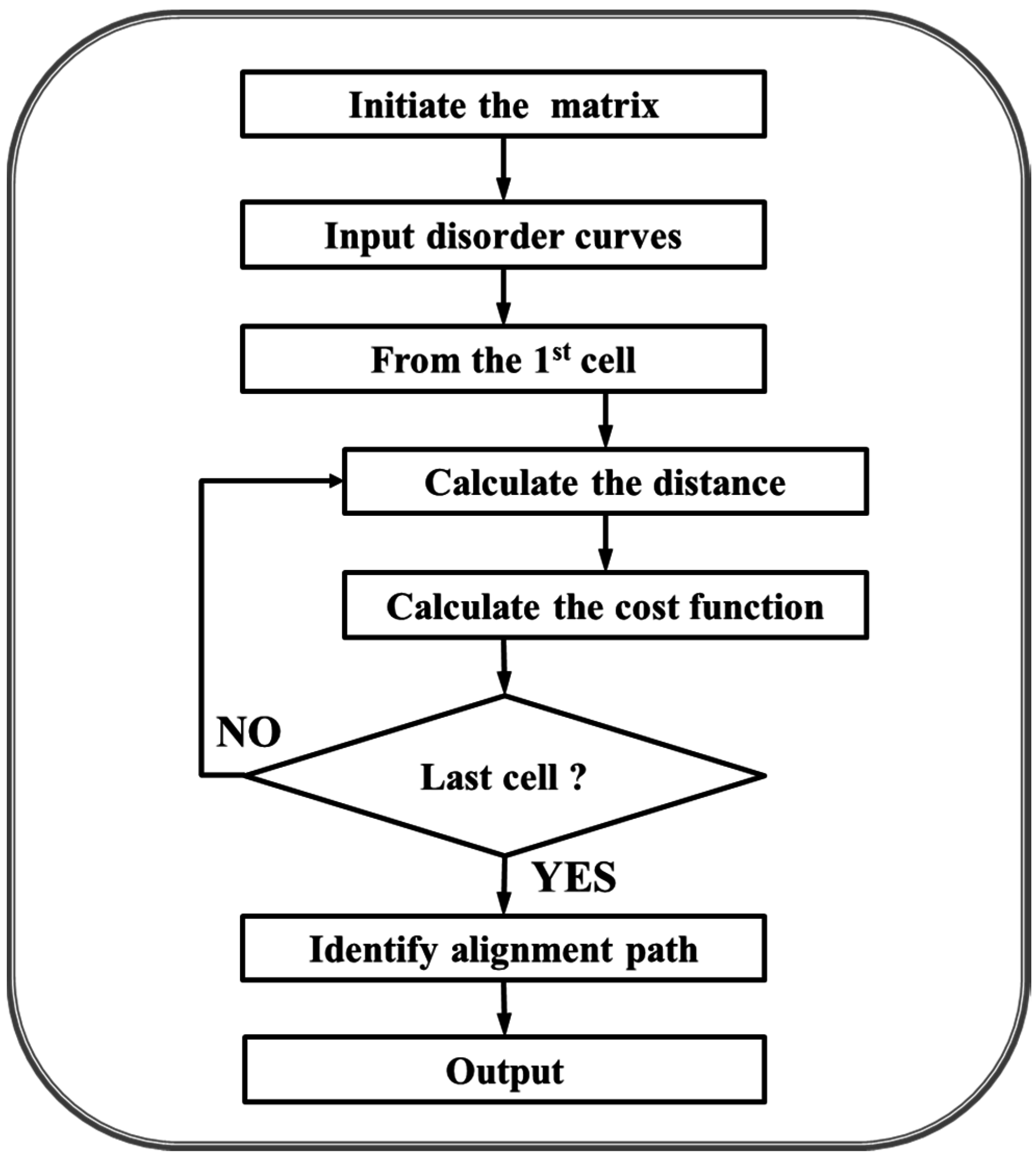
4.2. Dynamic Programming
4.3. Fraction of Matches
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Monastyrskyy, B.; Kryshtafovych, A.; Moult, J.; Tramontano, A.; Fidelis, K. Assessment of protein disorder region predictions in casp10. Proteins 2014, 82 (Suppl. 2), 127–137. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Urolagin, S.; Gurarslan, O.; Vihinen, M. Performance of protein disorder prediction programs on amino acid substitutions. Hum. Mutat. 2014, 35, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Simon, I.; Dosztanyi, Z. Prediction and analysis of intrinsically disordered proteins. Methods Mol. Biol. 2015, 1261, 35–59. [Google Scholar] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Adkins, J.N.; Lumb, K.J. Intrinsic structural disorder and sequence features of the cell cycle inhibitor p57kip2. Proteins 2002, 46, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Weathers, E.A.; Paulaitis, M.E.; Woolf, T.B.; Hoh, J.H. Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Lett. 2004, 576, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.C.; Lu, X.; Ross, E.D.; Woody, R.W. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 2006, 281, 1853–1856. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Yan, J.; Fan, X.; Mizianty, M.J.; Xue, B.; Wang, K.; Hu, G.; Uversky, V.N.; Kurgan, L. Exceptionally abundant exceptions: Comprehensive characterization of intrinsic disorder in all domains of life. Cell. Mol. Life Sci. 2015, 72, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Mizianty, M.J.; Kurgan, L. Genome-scale prediction of proteins with long intrinsically disordered regions. Proteins 2014, 82, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Uversky, V.N.; Xue, B. Local flexibility facilitates oxidization of buried methionine residues. Protein Pept. Lett. 2012, 19, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Williams, R.W.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Archaic chaos: Intrinsically disordered proteins in archaea. BMC Syst. Biol. 2010, 4 (Suppl. 1), S1. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic-Lazetic, G.M.; Mitic, N.S.; Kovacevic, J.J.; Obradovic, Z.; Malkov, S.N.; Beljanski, M.V. Bioinformatics analysis of disordered proteins in prokaryotes. BMC Bioinform. 2011, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Brown, C.J.; Obradovic, Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002, 62, 25–49. [Google Scholar] [PubMed]
- Minezaki, Y.; Homma, K.; Kinjo, A.R.; Nishikawa, K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006, 359, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 2007, 6, 1917–1932. [Google Scholar] [CrossRef] [PubMed]
- Eisenhaber, B.; Eisenhaber, F. Posttranslational modifications and subcellular localization signals: Indicators of sequence regions without inherent 3d structure? Curr. Protein Pept. Sci. 2007, 8, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Vacic, V.; Haynes, C.; Cocklin, R.R.; Mohan, A.; Heyen, J.W.; Goebl, M.G.; Iakoucheva, L.M. Identification, analysis, and prediction of protein ubiquitination sites. Proteins 2010, 78, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Edwards, Y.J.; Lobley, A.E.; Pentony, M.M.; Jones, D.T. Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Biol. 2009, 10, R50. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Mizianty, M.J.; Xue, B.; Kurgan, L.; Uversky, V.N. More than just tails: Intrinsic disorder in histone proteins. Mol. Biosyst. 2012, 8, 1886–1901. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Jeffers, V.; Sullivan, W.J.; Uversky, V.N. Protein intrinsic disorder in the acetylome of intracellular and extracellular toxoplasma gondii. Mol. Biosyst. 2013, 9, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Horikoshi, M. Histone acetylation: From code to web and router via intrinsically disordered regions. Curr. Pharm. Des. 2013, 19, 5019–5042. [Google Scholar] [CrossRef] [PubMed]
- Gough, J.; Dunker, A.K. Sequences and topology: Disorder, modularity, and post/pre translation modification. Curr. Opin. Struct. Biol. 2013, 23, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Chang, J.; Cheung, M.K.; Nong, W.; Li, L.; Lee, M.T.; Kwan, H.S. Human proteins with target sites of multiple post-translational modification types are more prone to be involved in disease. J. Proteome Res. 2014, 13, 2735–2748. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Hsu, W.L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014, 23, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Buljan, M.; Chalancon, G.; Dunker, A.K.; Bateman, A.; Balaji, S.; Fuxreiter, M.; Babu, M.M. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr. Opin. Struct. Biol. 2013, 23, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef] [PubMed]
- Pentony, M.M.; Jones, D.T. Modularity of intrinsic disorder in the human proteome. Proteins 2010, 78, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Trudeau, T.; Nassar, R.; Cumberworth, A.; Wong, E.T.; Woollard, G.; Gsponer, J. Structure and intrinsic disorder in protein autoinhibition. Structure 2013, 21, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Chu, X.; Hagen, S.J.; Han, W.; Wang, E. Multi-scaled explorations of binding-induced folding of intrinsically disordered protein inhibitor ia3 to its target enzyme. PLoS Comput. Biol. 2011, 7, e1001118. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Shah, S.P.; Gritsyna, Y.; Hitchcock-DeGregori, S.E.; Kostyukova, A.S. Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. J. Mol. Recognit. 2011, 24, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Vuzman, D.; Levy, Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol. Biosyst. 2012, 8, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Mittag, T.; Kay, L.E.; Forman-Kay, J.D. Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recognit. 2010, 23, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Bergantino, F.; Guariniello, S.; Raucci, R.; Colonna, G.; de Luca, A.; Normanno, N.; Costantini, S. Structure-fluctuation-function relationships of seven pro-angiogenic isoforms of vegfa, important mediators of tumorigenesis. Biochim. Biophys. Acta 2015, 1854, 410–425. [Google Scholar] [CrossRef] [PubMed]
- Mileo, E.; Lorenzi, M.; Erales, J.; Lignon, S.; Puppo, C.; Le Breton, N.; Etienne, E.; Marque, S.R.; Guigliarelli, B.; Gontero, B.; et al. Dynamics of the intrinsically disordered protein cp12 in its association with gapdh in the green alga chlamydomonas reinhardtii: A fuzzy complex. Mol. Biosyst. 2013, 9, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Vacic, V.; Oldfield, C.J.; Mohan, A.; Radivojac, P.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Characterization of molecular recognition features, morfs, and their binding partners. J. Proteome Res. 2007, 6, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef] [PubMed]
- Disfani, F.M.; Hsu, W.L.; Mizianty, M.J.; Oldfield, C.J.; Xue, B.; Dunker, A.K.; Uversky, V.N.; Kurgan, L. Morfpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics 2012, 28, i75–i83. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Noguchi, T.; Tominaga, D.; Yamana, H. Mfspssmpred: Identifying short disorder-to-order binding regions in disordered proteins based on contextual local evolutionary conservation. BMC Bioinform. 2013, 14, 300. [Google Scholar] [CrossRef] [PubMed]
- Malhis, N.; Gsponer, J. Computational identification of MoRFs in protein sequences. Bioinformatics 2015, 31, 1738–1744. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Retro-morfs: Identifying protein binding sites by normal and reverse alignment and intrinsic disorder prediction. Int. J. Mol. Sci. 2010, 11, 3725–3747. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: Intrinsically disordered n-terminal domains of gras proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 2011, 77, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Oldfield, C.J.; Van, Y.Y.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder and induced pluripotent stem cells. Mol. Biosyst. 2012, 8, 134–150. [Google Scholar] [CrossRef] [PubMed]
- Brunquell, J.; Yuan, J.; Erwin, A.; Westerheide, S.D.; Xue, B. Dbc1/ccar2 and ccar1 are largely disordered proteins that have evolved from one common ancestor. Biomed. Res. Int. 2014, 2014, 418458. [Google Scholar] [CrossRef] [PubMed]
- Lise, S.; Jones, D.T. Sequence patterns associated with disordered regions in proteins. Proteins 2005, 58, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.L.; Escouto, G.B.; Verli, H. Structural glycobiology of heparinase II from pedobacter heparinus. J. Biomol. Struct. Dyn. 2014, 32, 1092–1102. [Google Scholar] [CrossRef] [PubMed]
- Moroz, N.A.; Novak, S.M.; Azevedo, R.; Colpan, M.; Uversky, V.N.; Gregorio, C.C.; Kostyukova, A.S. Alteration of tropomyosin-binding properties of tropomodulin-1 affects its capping ability and localization in skeletal myocytes. J. Biol. Chem. 2013, 288, 4899–4907. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.; Rae, G.M.; Wu, R.M.; Walton, E.F.; Xue, B.; Hellens, R.P.; Uversky, V.N. Actinidia DRM1—An intrinsically disordered protein whose mrna expression is inversely correlated with spring budbreak in kiwifruit. PLoS ONE 2013, 8, e57354. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Greenwood, D.R.; Templeton, M.D.; Libich, D.S.; McGhie, T.K.; Xue, B.; Yoon, M.; Cui, W.; Kirk, C.A.; Jones, W.T.; et al. The intrinsically disordered structural platform of the plant defence hub protein rpm1-interacting protein 4 provides insights into its mode of action in the host-pathogen interface and evolution of the nitrate-induced domain protein family. FEBS J. 2014, 281, 3955–3979. [Google Scholar] [CrossRef] [PubMed]
- Redwan, E.M.; Xue, B.; Almehdar, H.A.; Uversky, V.N. Disorder in milk proteins: Caseins, intrinsically disordered colloids. Curr. Protein Pept. Sci. 2015, 16, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Almehdar, H.A.; El-Fakharany, E.M.; Uversky, V.N.; Redwan, E.M. Disorder in milk proteins: Structure, functional disorder, and biocidal potentials of lactoperoxidase. Curr. Protein Pept. Sci. 2015, 16, 352–365. [Google Scholar] [CrossRef] [PubMed]
- Albar, A.H.; Almehdar, H.A.; Uversky, V.N.; Redwan, E.M. Structural heterogeneity and multifunctionality of lactoferrin. Curr. Protein Pept. Sci. 2014, 15, 778–797. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Proteins without unique 3D structures: Biotechnological applications of intrinsically unstable/disordered proteins. Biotechnol. J. 2015, 10, 256–366. [Google Scholar] [CrossRef] [PubMed]
- Vintsyuk, T.K. Speech discrimination by dynamic programming. Kibernetika 1968, 4, 81–88. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Wagner, R.A.; Fischer, M.J. The string-to-string correction problem. J. ACM 1974, 21, 168–173. [Google Scholar] [CrossRef]
- Anderson, C.W.; Appella, E. Signaling to the p53 tumor suppressor through pathways activated by genotoxic and nongenotoxic stress. In Handbook of Cell Signaling; Bradshaw, R.A., Dennis, E.A., Eds.; Academic Press: New York, NY, USA, 2004; pp. 237–247. [Google Scholar]
- Joerger, A.C.; Fersht, A.R. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 2008, 77, 557–582. [Google Scholar] [CrossRef] [PubMed]
- Collavin, L.; Lunardi, A.; del Sal, G. P53-family proteins and their regulators: Hubs and spokes in tumor suppression. Cell Death Differ. 2010, 17, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.F.; Kaufman, M.H.; Harrison, D.J.; Clarke, A.R. High-frequency developmental abnormalities in p53-deficient mice. Curr. Biol. 1995, 5, 931–936. [Google Scholar] [CrossRef]
- Hollstein, M.; Sidransky, D.; Vogelstein, B.; Harris, C.C. P53 mutations in human cancers. Science 1991, 253, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Midic, U.; Xie, H.; Xue, B.; Vucetic, S.; Iakoucheva, L.M.; Obradovic, Z.; Dunker, A.K. Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genomics 2009, 10 (Suppl. 1), S7. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Brown, C.J.; Dunker, A.K.; Uversky, V.N. Intrinsically disordered regions of p53 family are highly diversified in evolution. Biochim. Biophys. Acta 2013, 1834, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Uversky, A.V.; Xue, B.; Peng, Z.; Kurgan, L.; Uversky, V.N. On the intrinsic disorder status of the major players in programmed cell death pathways. F1000Research 2013, 2, 190. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Xue, B.; Kurgan, L.; Uversky, V.N. Resilience of death: Intrinsic disorder in proteins involved in the programmed cell death. Cell Death Differ. 2013, 20, 1257–1267. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Meng, J.; Yang, J.Y.; Yang, M.Q.; Uversky, V.N.; Dunker, A.K. Flexible nets: Disorder and induced fit in the associations of p53 and 14–3-3 with their partners. BMC Genomics 2008, 9 (Suppl. 1), S1. [Google Scholar] [CrossRef] [PubMed]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D2p2: Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. Iupred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 2005, 61 (Suppl. 7), 176–182. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kinoshita, K. Prdos: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; di Domenico, T.; Tosatto, S.C. Espritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Osada, M.; Ohba, M.; Kawahara, C.; Ishioka, C.; Kanamaru, R.; Katoh, I.; Ikawa, Y.; Nimura, Y.; Nakagawara, A.; Obinata, M.; et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 1998, 4, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Kaghad, M.; Wang, Y.; Gillett, E.; Fleming, M.D.; Dotsch, V.; Andrews, N.C.; Caput, D.; McKeon, F. P63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 1998, 2, 305–316. [Google Scholar] [CrossRef]
- Kaghad, M.; Bonnet, H.; Yang, A.; Creancier, L.; Biscan, J.C.; Valent, A.; Minty, A.; Chalon, P.; Lelias, J.M.; Dumont, X.; et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997, 90, 809–819. [Google Scholar] [CrossRef]
- Yang, A.; Kaghad, M.; Caput, D.; McKeon, F. On the shoulders of giants: P63, p73 and the rise of p53. Trends Genet. 2002, 18, 90–95. [Google Scholar] [CrossRef]
- Murray-Zmijewski, F.; Lane, D.P.; Bourdon, J.C. P53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006, 13, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. Pondr-fit: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting intrinsic disorder in proteins: An overview. Cell Res. 2009, 19, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Sickmeier, M.; Hamilton, J.A.; LeGall, T.; Vacic, V.; Cortese, M.S.; Tantos, A.; Szabo, B.; Tompa, P.; Chen, J.; Uversky, V.N.; et al. Disprot: The database of disordered proteins. Nucleic Acids Res. 2007, 35, D786–D793. [Google Scholar] [CrossRef] [PubMed]
- Myers, C.S.; Rabiner, L.R. A comparative-study of several dynamic time-warping algorithms for connected-word recognition. Bell Syst. Tech. J. 1981, 60, 1389–1409. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrovich, A.; Borne, A.; Uversky, V.N.; Xue, B. Identifying Similar Patterns of Structural Flexibility in Proteins by Disorder Prediction and Dynamic Programming. Int. J. Mol. Sci. 2015, 16, 13829-13849. https://doi.org/10.3390/ijms160613829
Petrovich A, Borne A, Uversky VN, Xue B. Identifying Similar Patterns of Structural Flexibility in Proteins by Disorder Prediction and Dynamic Programming. International Journal of Molecular Sciences. 2015; 16(6):13829-13849. https://doi.org/10.3390/ijms160613829
Chicago/Turabian StylePetrovich, Aidan, Adam Borne, Vladimir N. Uversky, and Bin Xue. 2015. "Identifying Similar Patterns of Structural Flexibility in Proteins by Disorder Prediction and Dynamic Programming" International Journal of Molecular Sciences 16, no. 6: 13829-13849. https://doi.org/10.3390/ijms160613829







