A Structural Overview of RNA-Dependent RNA Polymerases from the Flaviviridae Family
Abstract
:1. Introduction
2. The Architecture of Flaviviridae RdRP Protein and Important Components for Polymerase Catalysis
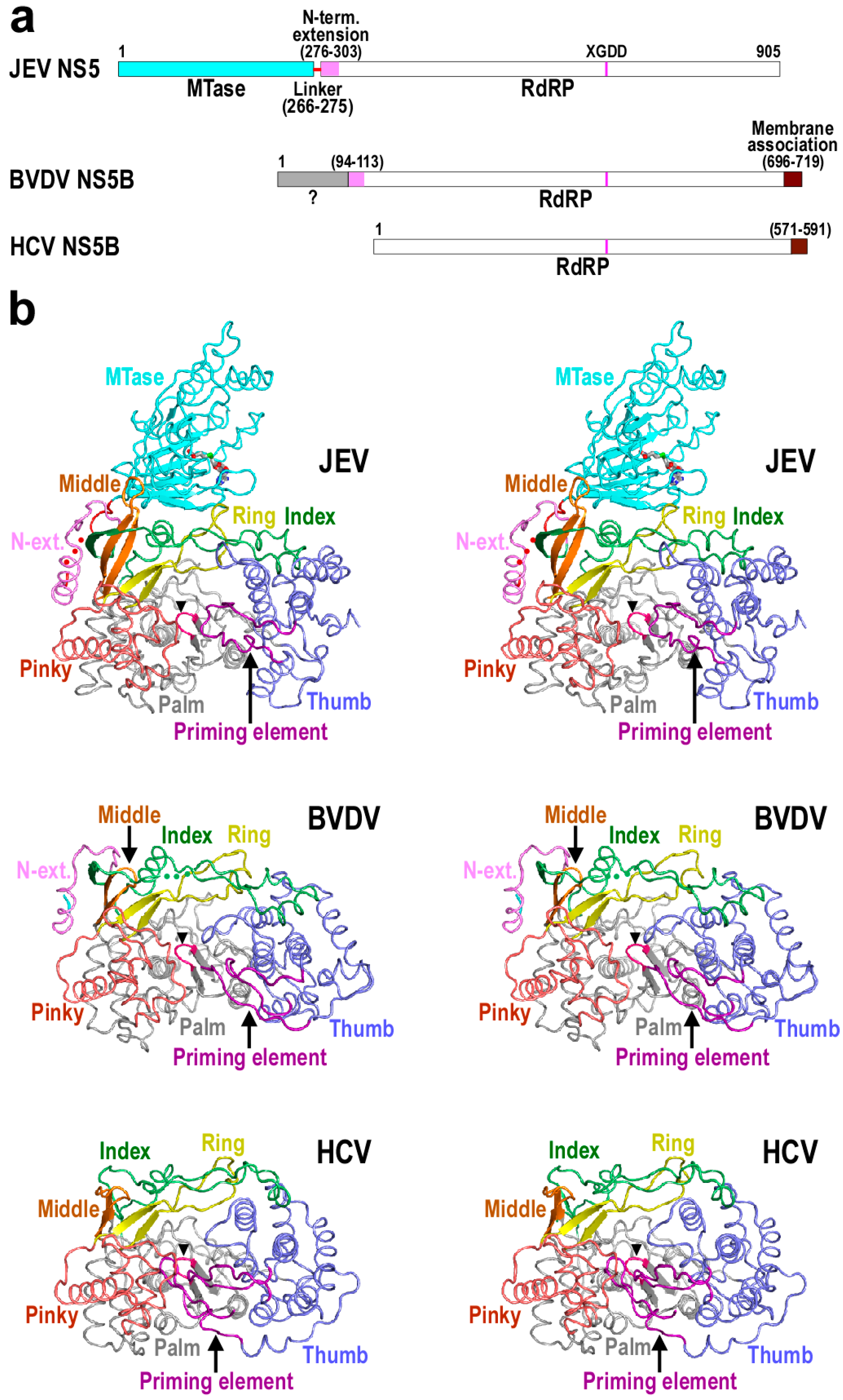

3. Structural Basis for Polymerase Catalysis
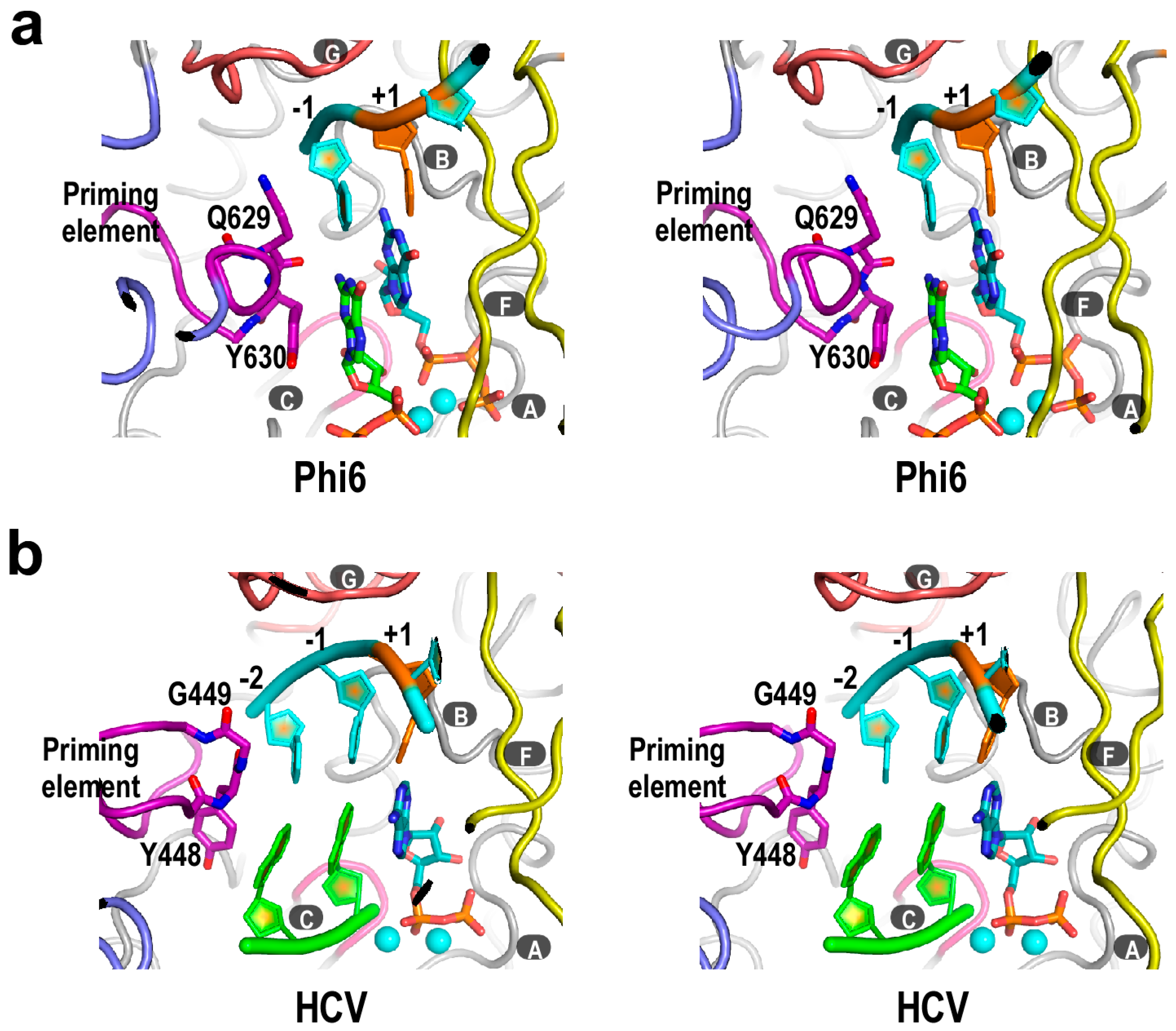
4. Flaviviridae RdRP Regulation in Cis
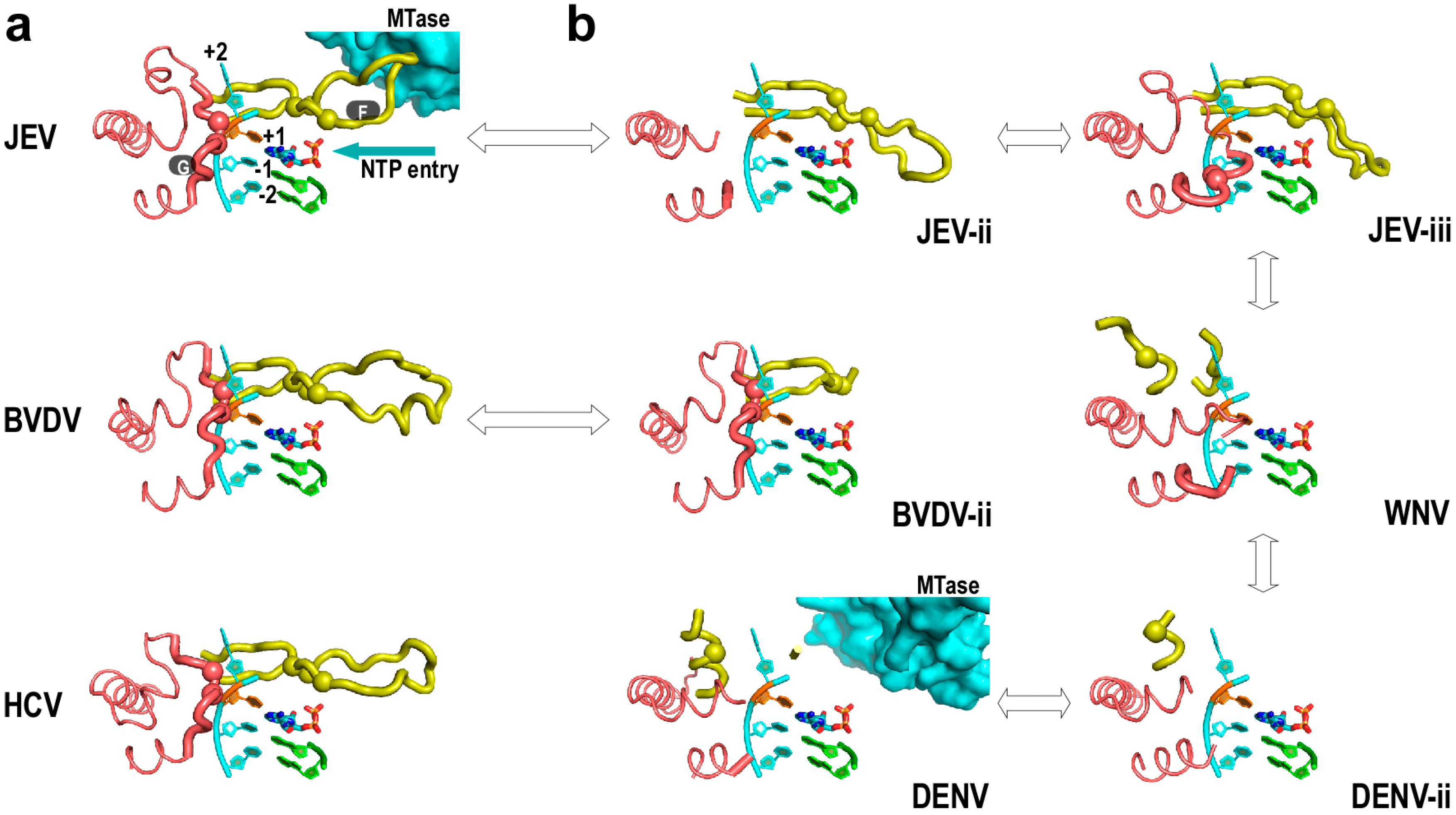
5. Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kato, N.; Hijikata, M.; Ootsuyama, Y.; Nakagawa, M.; Ohkoshi, S.; Sugimura, T.; Shimotohno, K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 1990, 87, 9524–9528. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Brinton, M.A.; Gaidamovich, S.; Horzinek, M.C.; Igarashi, A.; Kaariainen, L.; Lvov, D.K.; Porterfield, J.S.; Russell, P.K.; Trent, D.W. Flaviviridae . Intervirology 1985, 24, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Brinton, M.A.; Gaidamovich, S.; Horzinek, M.C.; Igarashi, A.; Kaariainen, L.; Lvov, D.K.; Porterfield, J.S.; Russell, P.K.; Trent, D.W. Togaviridae. Intervirology 1985, 24, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Tahara, S.M.; Lai, M.M. The 3ʹ-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 1998, 72, 8789–8796. [Google Scholar] [PubMed]
- Filomatori, C.V.; Lodeiro, M.F.; Alvarez, D.E.; Samsa, M.M.; Pietrasanta, L.; Gamarnik, A.V. A 5ʹ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006, 20, 2238–2249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Dong, H.; Zhou, Y.; Shi, P.Y. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5ʹ stem-loop of genomic RNA. J. Virol. 2008, 82, 7047–7058. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Lohmann, V.; Krieger, N.; Bartenschlager, R. Sequences in the 5ʹ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001, 75, 12047–12057. [Google Scholar] [CrossRef] [PubMed]
- Bisaillon, M.; Lemay, G. Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology 1997, 236, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tsukiyama-Kohara, K.; Iizuka, N.; Kohara, M.; Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992, 66, 1476–1483. [Google Scholar] [PubMed]
- Poole, T.L.; Wang, C.; Popp, R.A.; Potgieter, L.N.; Siddiqui, A.; Collett, M.S. Pestivirus translation initiation occurs by internal ribosome entry. Virology 1995, 206, 750–754. [Google Scholar] [CrossRef]
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Membranous replication factories induced by plus-strand RNA viruses. Viruses 2014, 6, 2826–2857. [Google Scholar] [CrossRef] [PubMed]
- Egloff, M.P.; Benarroch, D.; Selisko, B.; Romette, J.L.; Canard, B. An RNA cap (nucleoside-2'-O-)-methyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. EMBO J. 2002, 21, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- Malet, H.; Egloff, M.P.; Selisko, B.; Butcher, R.E.; Wright, P.J.; Roberts, M.; Gruez, A.; Sulzenbacher, G.; Vonrhein, C.; Bricogne, G.; et al. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 2007, 282, 10678–10689. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.L.; Xu, T.; Chen, Y.L.; Malet, H.; Egloff, M.P.; Canard, B.; Vasudevan, S.G.; Lescar, J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 2007, 81, 4753–4765. [Google Scholar] [CrossRef] [PubMed]
- Geiss, B.J.; Thompson, A.A.; Andrews, A.J.; Sons, R.L.; Gari, H.H.; Keenan, S.M.; Peersen, O.B. Analysis of flavivirus NS5 methyltransferase cap binding. J. Mol. Biol. 2009, 385, 1643–1654. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Gong, P. Crystal structure of the full-length Japanese Encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013, 9, e1003549. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Soh, T.S.; Zheng, J.; Chan, K.W.; Phoo, W.W.; Lee, C.C.; Tay, M.Y.; Swaminathan, K.; Cornvik, T.C.; Lim, S.P.; et al. A crystal structure of the dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog. 2015, 11, e1004682. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lu, G.; Zhang, B.; Gong, P. Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J. Virol. 2015, 89, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Shan, C.; Deng, C.L.; Ye, H.Q.; Shi, P.Y.; Yuan, Z.M.; Gong, P.; Zhang, B. The interface between methyltransferase and polymerase of NS5 is essential for flavivirus replication. PLoS Negl. Trop. Dis. 2014, 8, e2891. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.H.; Gallei, A.; Becher, P.; Rossmann, M.G. The structure of bovine viral diarrhea virus RNA-dependent RNA polymerase and its amino-terminal domain. Structure 2006, 14, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Theobald, D.L.; Wuttke, D.S. THESEUS: Maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics 2006, 22, 2171–2172. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.A.; Peersen, O.B. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004, 23, 3462–3471. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.W.; Steitz, T.A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 2004, 116, 393–404. [Google Scholar] [CrossRef]
- Li, Y.; Korolev, S.; Waksman, G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998, 17, 7514–7525. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Peersen, O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 2010, 107, 22505–22510. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.H.; Kao, C.C. Characterization of RNA products associated with or aborted by a viral RNA-dependent RNA polymerase. Virology 1997, 236, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Butcher, S.J.; Grimes, J.M.; Makeyev, E.V.; Bamford, D.H.; Stuart, D.I. A mechanism for initiating RNA-dependent RNA polymerization. Nature 2001, 410, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, A.; Detjen, B.; Pozzatti, R.; Wimmer, E. The location of the polio genome protein in viral RNAs and its implication for RNA synthesis. Nature 1977, 268, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Pringle, F.M.; Zeddam, J.L.; Luke, B.T.; Cameron, C.E.; Kalmakoff, J.; Hanzlik, T.N.; Gordon, K.H.; Ward, V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002, 324, 47–62. [Google Scholar] [CrossRef]
- Poch, O.; Sauvaget, I.; Delarue, M.; Tordo, N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989, 8, 3867–3874. [Google Scholar] [PubMed]
- Beese, L.S.; Steitz, T.A. Structural basis for the 3ʹ–5ʹ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991, 10, 25–33. [Google Scholar] [PubMed]
- Crotty, S.; Gohara, D.; Gilligan, D.K.; Karelsky, S.; Cameron, C.E.; Andino, R. Manganese-dependent polioviruses caused by mutations within the viral polymerase. J. Virol. 2003, 77, 5378–5388. [Google Scholar] [CrossRef] [PubMed]
- Sholders, A.J.; Peersen, O.B. Distinct conformations of a putative translocation element in poliovirus polymerase. J. Mol. Biol. 2014, 426, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.; Smidansky, E.D.; Arnold, J.J.; Maksimchuk, K.R.; Moustafa, I.; Uchida, A.; Gotte, M.; Konigsberg, W.; Cameron, C.E. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009, 16, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, D.; Tomita, K. Molecular basis for RNA polymerization by Qbeta replicase. Nat. Struct. Mol. Biol. 2012, 19, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Vakharia, V.N.; Tao, Y.J. The structure of a birnavirus polymerase reveals a distinct active site topology. Proc. Natl. Acad. Sci. USA 2007, 104, 7385–7390. [Google Scholar] [CrossRef] [PubMed]
- Reich, S.; Guilligay, D.; Pflug, A.; Malet, H.; Berger, I.; Crepin, T.; Hart, D.; Lunardi, T.; Nanao, M.; Ruigrok, R.W.; et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 2014, 516, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chopra, R.; Verdine, G.L.; Harrison, S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science 1998, 282, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Gillis, A.J.; Schuller, A.P.; Skordalakes, E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 2008, 455, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Bruenn, J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003, 31, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Appleby, T.C.; Perry, J.K.; Murakami, E.; Barauskas, O.; Feng, J.; Cho, A.; Fox, D., 3rd; Wetmore, D.R.; McGrath, M.E.; Ray, A.S.; et al. Viral replication. structural basis for RNA replication by the hepatitis C virus polymerase. Science 2015, 347, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, N.G.; Filomatori, C.V.; Gamarnik, A.V. The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J. Virol. 2011, 85, 5745–5756. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Kortus, M.G.; Nix, J.C.; Davis, R.E.; Peersen, O.B. Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PLoS ONE 2013, 8, e60272. [Google Scholar] [CrossRef] [PubMed]
- Zamyatkin, D.F.; Parra, F.; Alonso, J.M.; Harki, D.A.; Peterson, B.R.; Grochulski, P.; Ng, K.K. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 2008, 283, 7705–7712. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Orta, C.; Arias, A.; Perez-Luque, R.; Escarmis, C.; Domingo, E.; Verdaguer, N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 2004, 279, 47212–47221. [Google Scholar] [CrossRef] [PubMed]
- Carpousis, A.J.; Gralla, J.D. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J. Mol. Biol. 1985, 183, 165–177. [Google Scholar] [CrossRef]
- Martin, C.T.; Muller, D.K.; Coleman, J.E. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry 1988, 27, 3966–3974. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Martin, C.T. Mechanism of instability in abortive cycling by T7 RNA polymerase. J. Biol. Chem. 2006, 281, 23533–23544. [Google Scholar] [CrossRef] [PubMed]
- Lesburg, C.A.; Cable, M.B.; Ferrari, E.; Hong, Z.; Mannarino, A.F.; Weber, P.C. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 1999, 6, 937–943. [Google Scholar] [PubMed]
- Hong, Z.; Cameron, C.E.; Walker, M.P.; Castro, C.; Yao, N.; Lau, J.Y.; Zhong, W. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 2001, 285, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Cherry, A.L.; Dennis, C.A.; Baron, A.; Eisele, L.E.; Thommes, P.A.; Jaeger, J. Hydrophobic and charged residues in the C-terminal arm of hepatitis C virus RNA-dependent RNA polymerase regulate initiation and elongation. J. Virol. 2015, 89, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Selisko, B.; Potisopon, S.; Agred, R.; Priet, S.; Varlet, I.; Thillier, Y.; Sallamand, C.; Debart, F.; Vasseur, J.J.; Canard, B. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 2012, 8, e1002912. [Google Scholar] [CrossRef] [PubMed]
- Takeda, N.; Kuhn, R.J.; Yang, C.F.; Takegami, T.; Wimmer, E. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J. Virol. 1986, 60, 43–53. [Google Scholar] [PubMed]
- Paul, A.V.; Wimmer, E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Orta, C.; Arias, A.; Agudo, R.; Perez-Luque, R.; Escarmis, C.; Domingo, E.; Verdaguer, N. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 2006, 25, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Gruez, A.; Selisko, B.; Roberts, M.; Bricogne, G.; Bussetta, C.; Jabafi, I.; Coutard, B.; de Palma, A.M.; Neyts, J.; Canard, B. The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J. Virol. 2008, 82, 9577–9590. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, Y.; Shan, C.; Sun, Y.; Xu, P.; Zhou, H.; Yang, C.; Shi, P.Y.; Rao, Z.; Zhang, B.; et al. Crystal structure of enterovirus 71 RNA-dependent RNA polymerase complexed with its protein primer VPg: Implication for a trans mechanism of VPg uridylylation. J. Virol. 2013, 87, 5755–5768. [Google Scholar] [CrossRef] [PubMed]
- Mosley, R.T.; Edwards, T.E.; Murakami, E.; Lam, A.M.; Grice, R.L.; Du, J.; Sofia, M.J.; Furman, P.A.; Otto, M.J. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 2012, 86, 6503–6511. [Google Scholar] [CrossRef] [PubMed]
- Surana, P.; Satchidanandam, V.; Nair, D.T. RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state. Nucleic Acids Res. 2014, 42, 2758–2773. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.P.; Koh, J.H.; Seh, C.C.; Liew, C.W.; Davidson, A.D.; Chua, L.S.; Chandrasekaran, R.; Cornvik, T.C.; Shi, P.Y.; Lescar, J. A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J. Biol. Chem. 2013, 288, 31105–31114. [Google Scholar] [CrossRef] [PubMed]
- Potisopon, S.; Priet, S.; Collet, A.; Decroly, E.; Canard, B.; Selisko, B. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res. 2014, 42, 11642–11656. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.G.; Lim, S.P.; Chen, Y.L.; Liew, C.W.; Yap, L.; Lescar, J.; Shi, P.Y. Conformational Flexibility of the dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 2013, 87, 5291–5295. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.H.; Groarke, J.M.; Young, D.C.; Rossmann, M.G.; Pevear, D.C.; Kuhn, R.J.; Smith, J.L. Design, expression, and purification of a Flaviviridae polymerase using a high-throughput approach to facilitate crystal structure determination. Protein Sci. 2004, 13, 2685–2692. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Li, H.; Wang, Y.; Wang, X.; Wang, W.; Peng, J.; Chen, J.; Li, B. Characterization of the N-terminal domain of classical swine fever virus RNA-dependent RNA polymerase. J. Gen. Virol. 2006, 87, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Lai, V.C.; Kao, C.C.; Ferrari, E.; Park, J.; Uss, A.S.; Wright-Minogue, J.; Hong, Z.; Lau, J.Y. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 1999, 73, 10129–10136. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Liu, W.; Gong, P. A Structural Overview of RNA-Dependent RNA Polymerases from the Flaviviridae Family. Int. J. Mol. Sci. 2015, 16, 12943-12957. https://doi.org/10.3390/ijms160612943
Wu J, Liu W, Gong P. A Structural Overview of RNA-Dependent RNA Polymerases from the Flaviviridae Family. International Journal of Molecular Sciences. 2015; 16(6):12943-12957. https://doi.org/10.3390/ijms160612943
Chicago/Turabian StyleWu, Jiqin, Weichi Liu, and Peng Gong. 2015. "A Structural Overview of RNA-Dependent RNA Polymerases from the Flaviviridae Family" International Journal of Molecular Sciences 16, no. 6: 12943-12957. https://doi.org/10.3390/ijms160612943




