Micro RNA-124a Regulates Lipolysis via Adipose Triglyceride Lipase and Comparative Gene Identification 58
Abstract
:1. Introduction
1.1. Lipases and Lipolysis
1.2. Regulation of Lipases by miRNAs
2. Results
2.1. miR-124a Regulates Expression of Adipose Triglyceride Lipase (ATGL) and Comparative Gene Identification 58
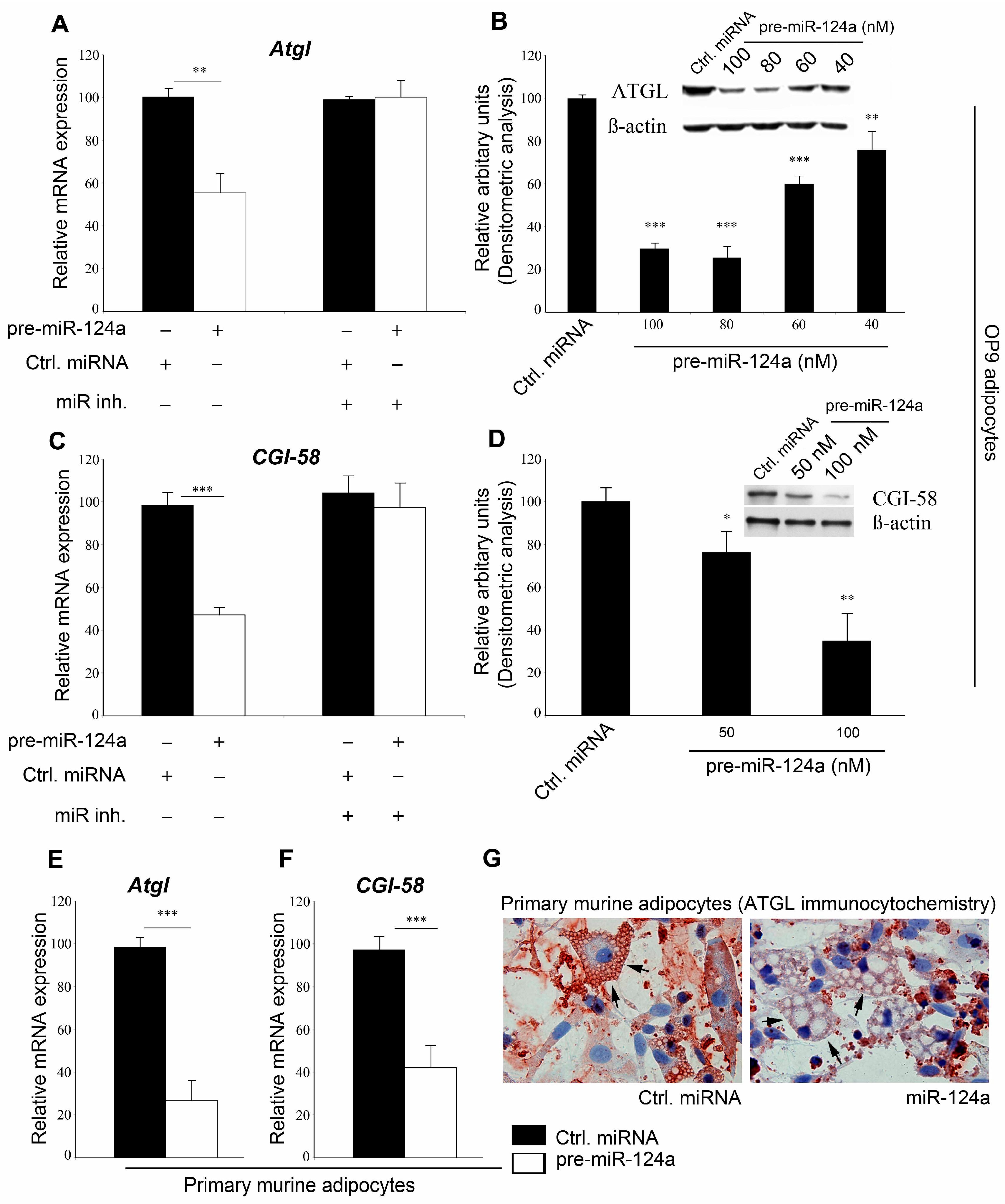
2.2. miR-124a Regulates ATGL and CGI-58 via Single Targeting Sequences in Their 3'UTRs
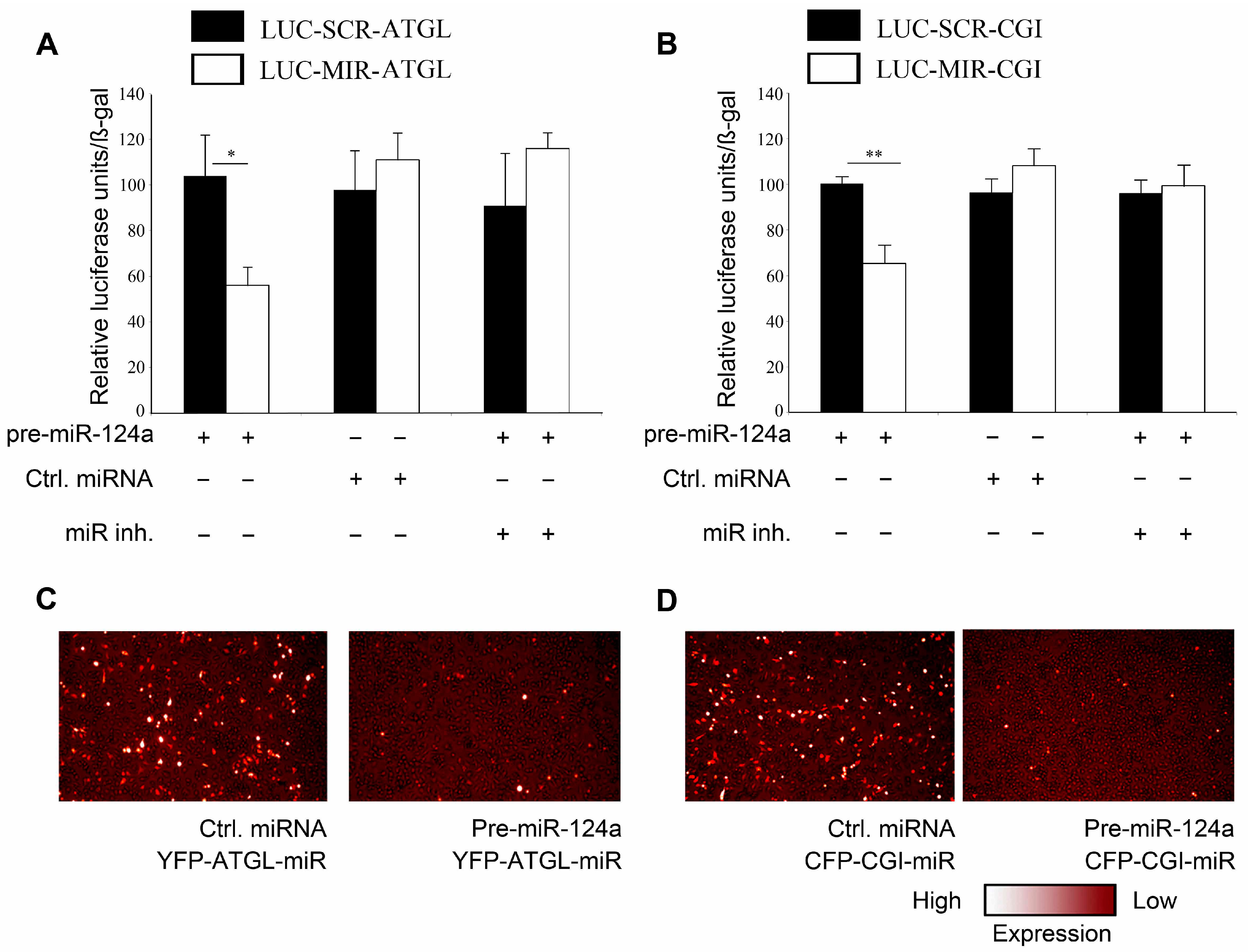
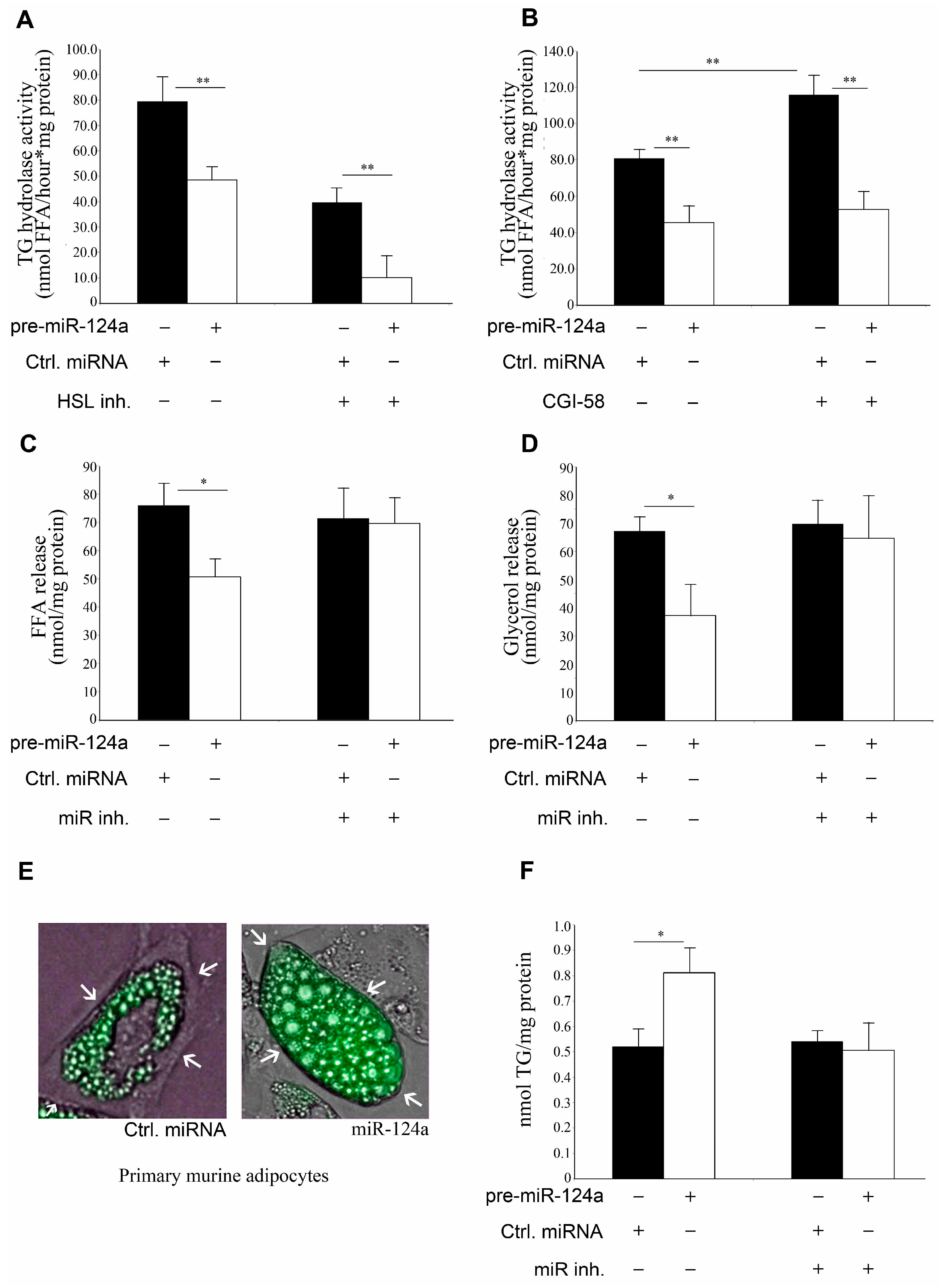
2.3. miR-124a Down-Regulates Lipolysis
2.4. Negative Correlation of miR-124a and Atgl Expression in Various Tissues

2.5. miR-124a Regulates the Expression of Atgl and Cgi-58 during Fasting Conditions
3. Experimental Section
3.1. Animal Care and Tissue Harvesting
3.2. Cell Lines and Constructs
3.4. Triacylgycerol Hydrolase Assay
3.5. Free Fatty Acid and Glycerol Release Assay
3.6. Intracellular TG Measurement
3.7. Microscopic Analysis of Adipocytes
3.8. Flow Cytometry
3.9. Luciferase Assay
3.10. Western Blot and Immunohistochemistry
3.11. qRT-PCR
3.12. Statistical Methods
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lass, A.; Zimmermann, R.; Oberer, M.; Zechner, R. Lipolysis—A highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog. Lipid Res. 2011, 50, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Wang, Y.; Sul, H.S. Lipolysis in adipocytes. Int. J. Biochem. Cell. Biol. 2010, 42, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Lass, A.; Zimmermann, R.; Haemmerle, G.; Riederer, M.; Schoiswohl, G.; Schweiger, M.; Kienesberger, P.; Strauss, J.G.; Gorkiewicz, G.; Zechner, R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in chanarin-dorfman syndrome. Cell. Metab. 2006, 3, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Omatsu, N.; Morimoto, E.; Nakashima, H.; Ueno, K.; Tanaka, T.; Satouchi, K.; Hirose, F.; Osumi, T. CGI-58 facilitates lipolysis on lipid droplets but is not involved in the vesiculation of lipid droplets caused by hormonal stimulation. J. Lipid Res. 2007, 48, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Lefevre, C.; Morava, E.; Mussini, J.M.; Laforet, P.; Negre-Salvayre, A.; Lathrop, M.; Salvayre, R. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat. Genet. 2007, 39, 28–30. [Google Scholar] [CrossRef] [PubMed]
- Haemmerle, G.; Lass, A.; Zimmermann, R.; Gorkiewicz, G.; Meyer, C.; Rozman, J.; Heldmaier, G.; Maier, R.; Theussl, C.; Eder, S.; et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 2006, 312, 734–737. [Google Scholar]
- Radner, F.P.; Streith, I.E.; Schoiswohl, G.; Schweiger, M.; Kumari, M.; Eichmann, T.O.; Rechberger, G.; Koefeler, H.C.; Eder, S.; Schauer, S.; et al. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58). J. Biol. Chem. 2010, 285, 7300–7311. [Google Scholar]
- Schweiger, M.; Lass, A.; Zimmermann, R.; Eichmann, T.O.; Zechner, R. Neutral lipid storage disease: Genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/abhd5. Am. J. Physiol.-Endocrinol. Metab. 2009, 297, E289–E296. [Google Scholar] [CrossRef] [PubMed]
- Albert, J.S.; Yerges-Armstrong, L.M.; Horenstein, R.B.; Pollin, T.I.; Sreenivasan, U.T.; Chai, S.; Blaner, W.S.; Snitker, S.; O’Connell, J.R.; Gong, D.W.; et al. Null mutation in hormone-sensitive lipase gene and risk of type 2 diabetes. N. Eng. J. Med. 2014, 370, 2307–2315. [Google Scholar]
- Taschler, U.; Radner, F.P.; Heier, C.; Schreiber, R.; Schweiger, M.; Schoiswohl, G.; Preiss-Landl, K.; Jaeger, D.; Reiter, B.; Koefeler, H.C.; et al. Monoglyceride lipase deficiency in mice impairs lipolysis and attenuates diet-induced insulin resistance. J. Biol. Chem. 2011, 286, 17467–17477. [Google Scholar]
- Haemmerle, G.; Zimmermann, R.; Strauss, J.G.; Kratky, D.; Riederer, M.; Knipping, G.; Zechner, R. Hormone-sensitive lipase deficiency in mice changes the plasma lipid profile by affecting the tissue-specific expression pattern of lipoprotein lipase in adipose tissue and muscle. J. Biol. Chem. 2002, 277, 12946–12952. [Google Scholar] [CrossRef] [PubMed]
- Osuga, J.; Ishibashi, S.; Oka, T.; Yagyu, H.; Tozawa, R.; Fujimoto, A.; Shionoiri, F.; Yahagi, N.; Kraemer, F.B.; Tsutsumi, O.; et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl. Acad. Sci. USA 2000, 97, 787–792. [Google Scholar]
- Buchebner, M.; Pfeifer, T.; Rathke, N.; Chandak, P.G.; Lass, A.; Schreiber, R.; Kratzer, A.; Zimmermann, R.; Sattler, W.; Koefeler, H.; et al. Cholesteryl ester hydrolase activity is abolished in HSL macrophages but unchanged in macrophages lacking KIAA1363. J. Lipid Res. 2010, 51, 2896–2908. [Google Scholar]
- Das, S.K.; Eder, S.; Schauer, S.; Diwoky, C.; Temmel, H.; Guertl, B.; Gorkiewicz, G.; Tamilarasan, K.P.; Kumari, P.; Trauner, M.; et al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science 2011, 333, 233–238. [Google Scholar]
- Parra, P.; Serra, F.; Palou, A. Expression of adipose micrornas is sensitive to dietary conjugated linoleic acid treatment in mice. PLoS ONE 2010, 5, e13005. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 2007, 27, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, E.D.; Veremeyko, T.; Barteneva, N.; Krichevsky, A.M.; Weiner, H.L. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 2011, 17, 64–70. [Google Scholar] [CrossRef]
- Rajasethupathy, P.; Fiumara, F.; Sheridan, R.; Betel, D.; Puthanveettil, S.V.; Russo, J.J.; Sander, C.; Tuschl, T.; Kandel, E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 2009, 63, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Kruger, J.; Rehmsmeier, M. Rnahybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2.3, 2–3. [Google Scholar]
- Wolins, N.E.; Quaynor, B.K.; Skinner, J.R.; Tzekov, A.; Park, C.; Choi, K.; Bickel, P.E. OP9 mouse stromal cells rapidly differentiate into adipocytes: Characterization of a useful new model of adipogenesis. J. Lipid Res. 2006, 47, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, M.; Schreiber, R.; Haemmerle, G.; Lass, A.; Fledelius, C.; Jacobsen, P.; Tornqvist, H.; Zechner, R.; Zimmermann, R. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 2006, 281, 40236–40241. [Google Scholar] [CrossRef] [PubMed]
- Fernyhough, M.E.; Vierck, J.L.; Hausman, G.J.; Mir, P.S.; Okine, E.K.; Dodson, M.V. Primary adipocyte culture: Adipocyte purification methods may lead to a new understanding of adipose tissue growth and development. Cytotechnology 2004, 46, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.S.; Jessen, N.; Jorgensen, J.O.; Moller, N.; Lund, S. Dissecting adipose tissue lipolysis: Molecular regulation and implications for metabolic disease. J. Mol. Endocrinol. 2014, 52, R199–R222. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.Y.; Chou, C.F.; Giovarelli, M.; Briata, P.; Gherzi, R.; Chen, C.Y. KSRP and microRNA 145 are negative regulators of lipolysis in white adipose tissue. Mol. Cell. Biol. 2014, 34, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, P.; Kandror, K.V. Adipose triglyceride lipase: A new target in the regulation of lipolysis by insulin. Curr. Diabetes Rev. 2011, 7, 270–277. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, S.K.; Stadelmeyer, E.; Schauer, S.; Schwarz, A.; Strohmaier, H.; Claudel, T.; Zechner, R.; Hoefler, G.; Vesely, P.W. Micro RNA-124a Regulates Lipolysis via Adipose Triglyceride Lipase and Comparative Gene Identification 58. Int. J. Mol. Sci. 2015, 16, 8555-8568. https://doi.org/10.3390/ijms16048555
Das SK, Stadelmeyer E, Schauer S, Schwarz A, Strohmaier H, Claudel T, Zechner R, Hoefler G, Vesely PW. Micro RNA-124a Regulates Lipolysis via Adipose Triglyceride Lipase and Comparative Gene Identification 58. International Journal of Molecular Sciences. 2015; 16(4):8555-8568. https://doi.org/10.3390/ijms16048555
Chicago/Turabian StyleDas, Suman K., Elke Stadelmeyer, Silvia Schauer, Anna Schwarz, Heimo Strohmaier, Thiery Claudel, Rudolf Zechner, Gerald Hoefler, and Paul W. Vesely. 2015. "Micro RNA-124a Regulates Lipolysis via Adipose Triglyceride Lipase and Comparative Gene Identification 58" International Journal of Molecular Sciences 16, no. 4: 8555-8568. https://doi.org/10.3390/ijms16048555





