The Neuronal-Specific SGK1.1 (SGK1_v2) Kinase as a Transcriptional Modulator of BAG4, Brox, and PPP1CB Genes Expression
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening the Subtracted cDNA Library Identified Six Independent and Differentially Expressed Clones
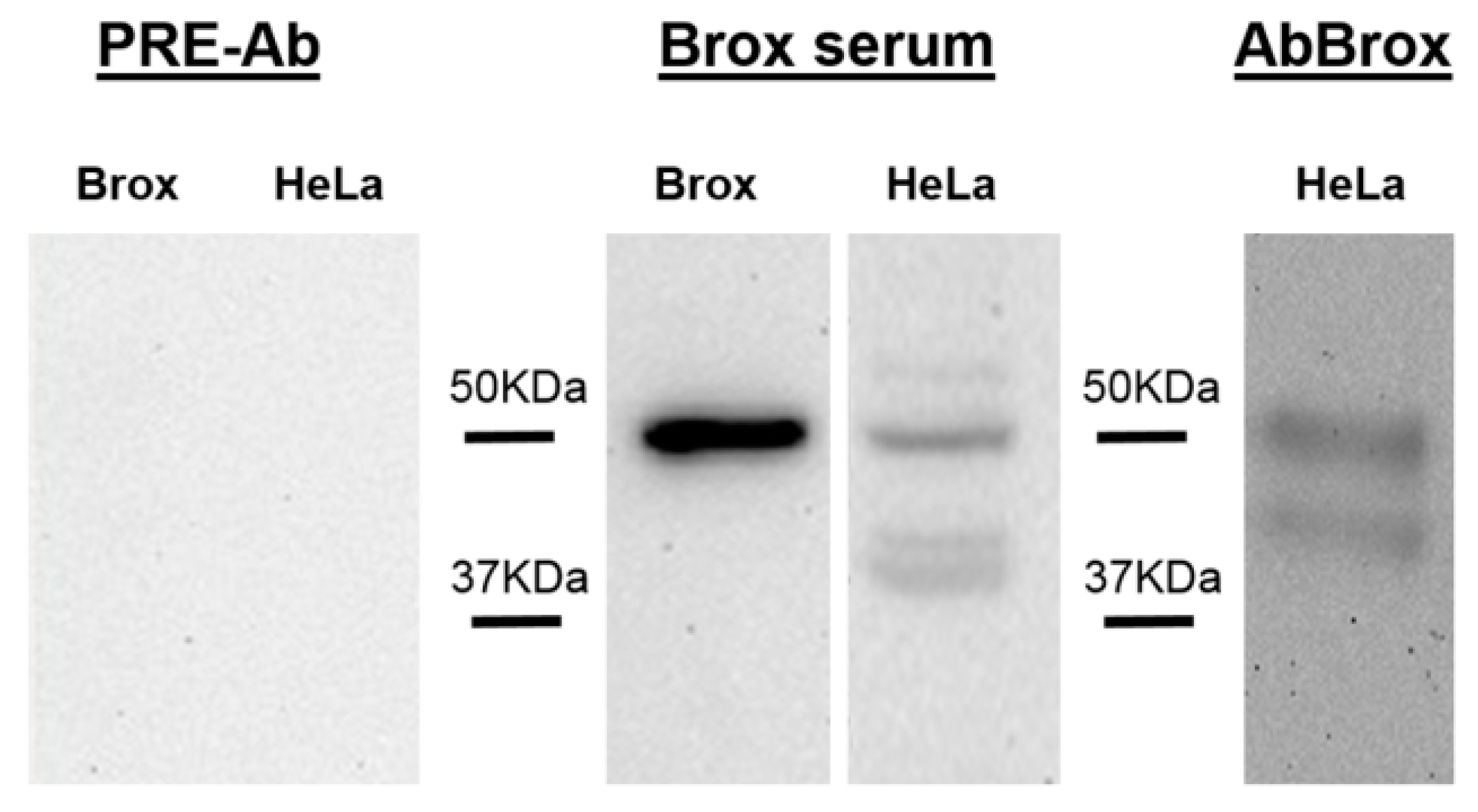
2.2. Anti-Brox Antibody (Brox-Ab) Specificity
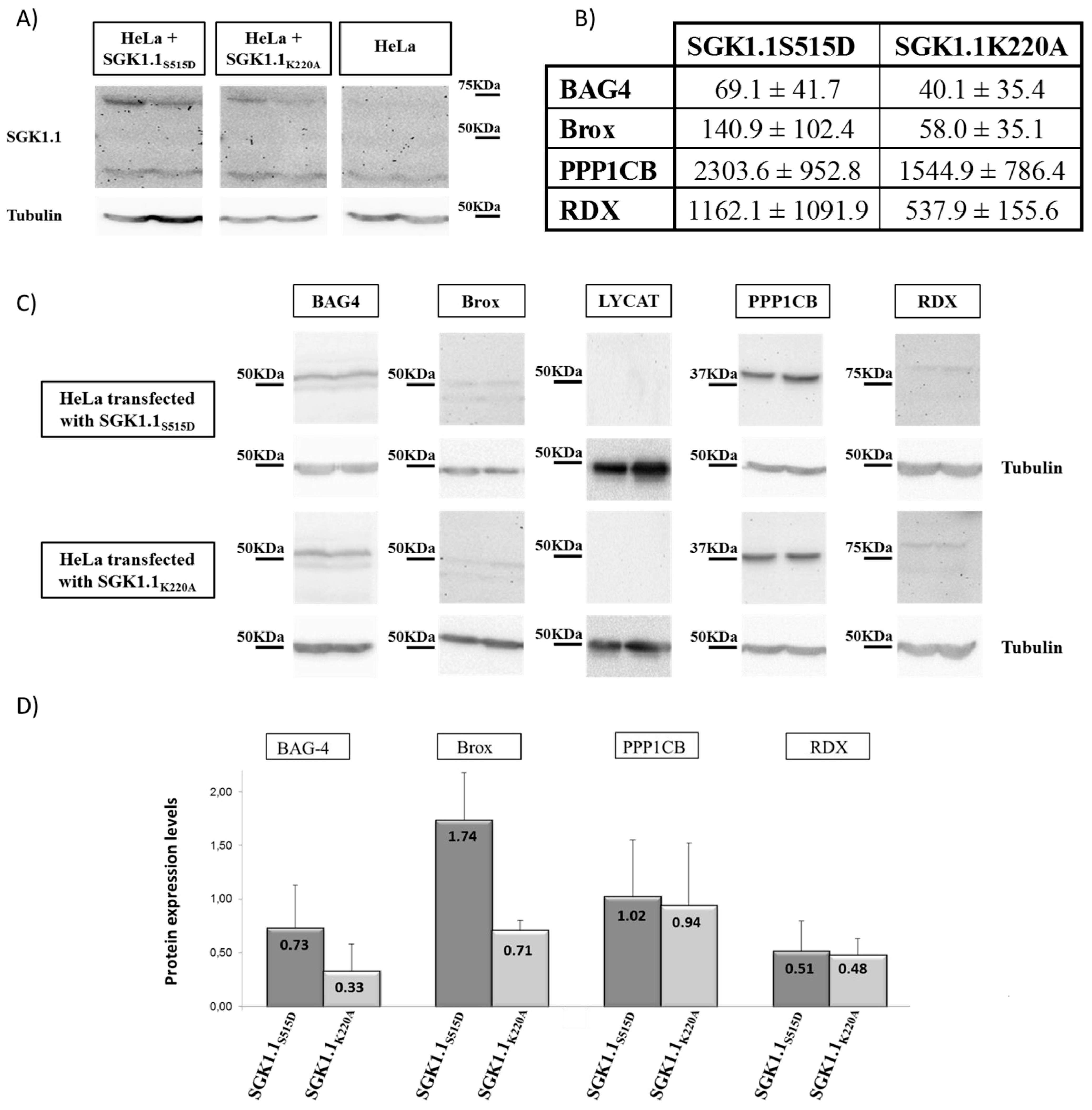
2.3. Validation of Genes Identified by Subtraction Screening
| Gene | SGK1.1S515D | SGK1.1K220A | p Value |
|---|---|---|---|
| BAG4 | 96.5 ± 62.1 | 41.3 ± 27.3 | 0.001 |
| Brox | 178.8 ± 101.9 | 81.7 ± 61.6 | 0.001 |
| LYCAT | 319.9 ± 210.1 | 196.6 ± 117.4 | 0.029 |
| PPP1CB | 2492.9 ± 1625.4 | 1528.0 ± 832.2 | 0.023 |
| RDX | 1355.0 ± 961.6 | 829.7 ± 638.5 | 0.042 |
| C6ORF62 | 4684.2 ± 2856.7 | 4341.8 ± 3026.0 | 0.715 |
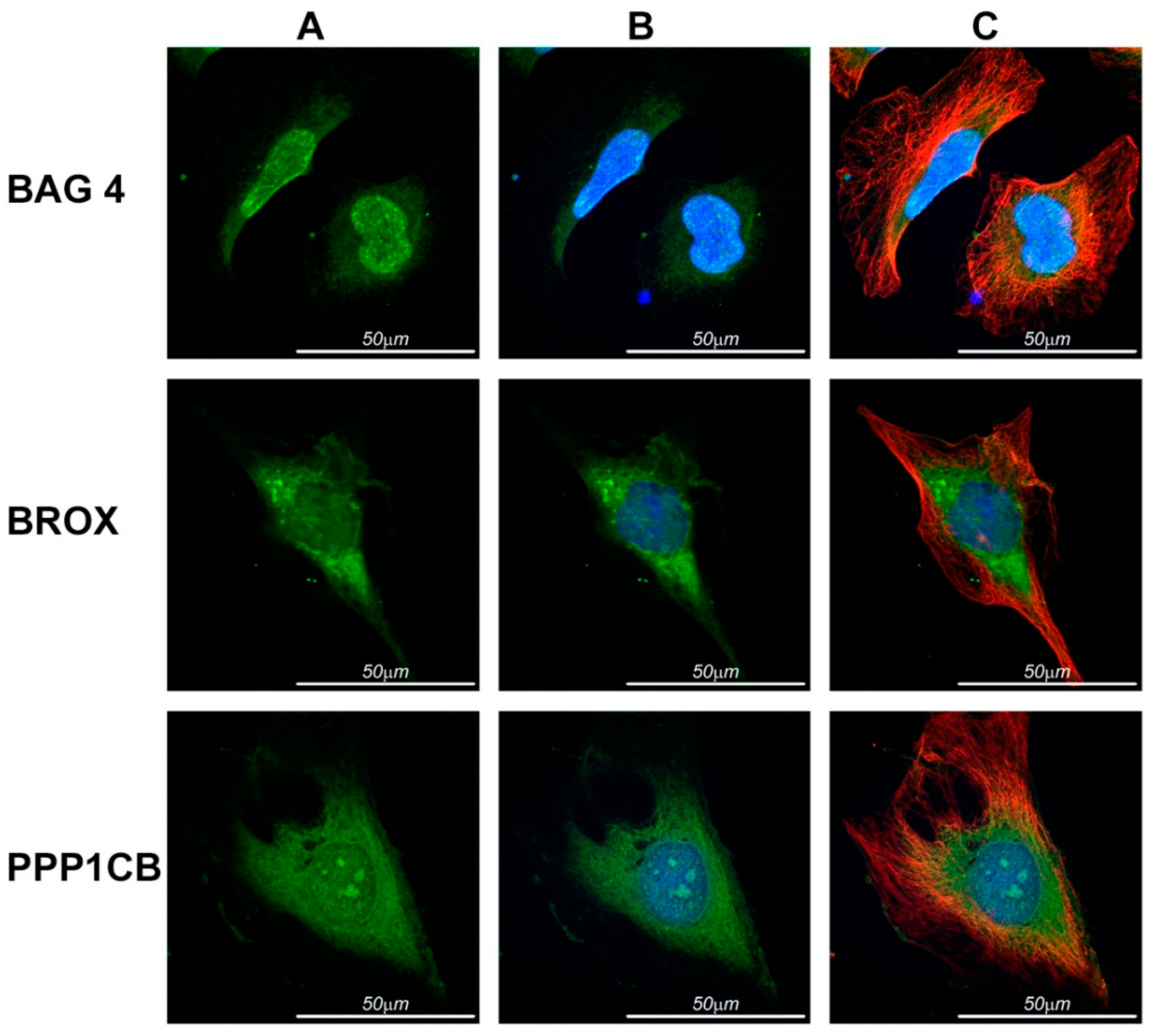
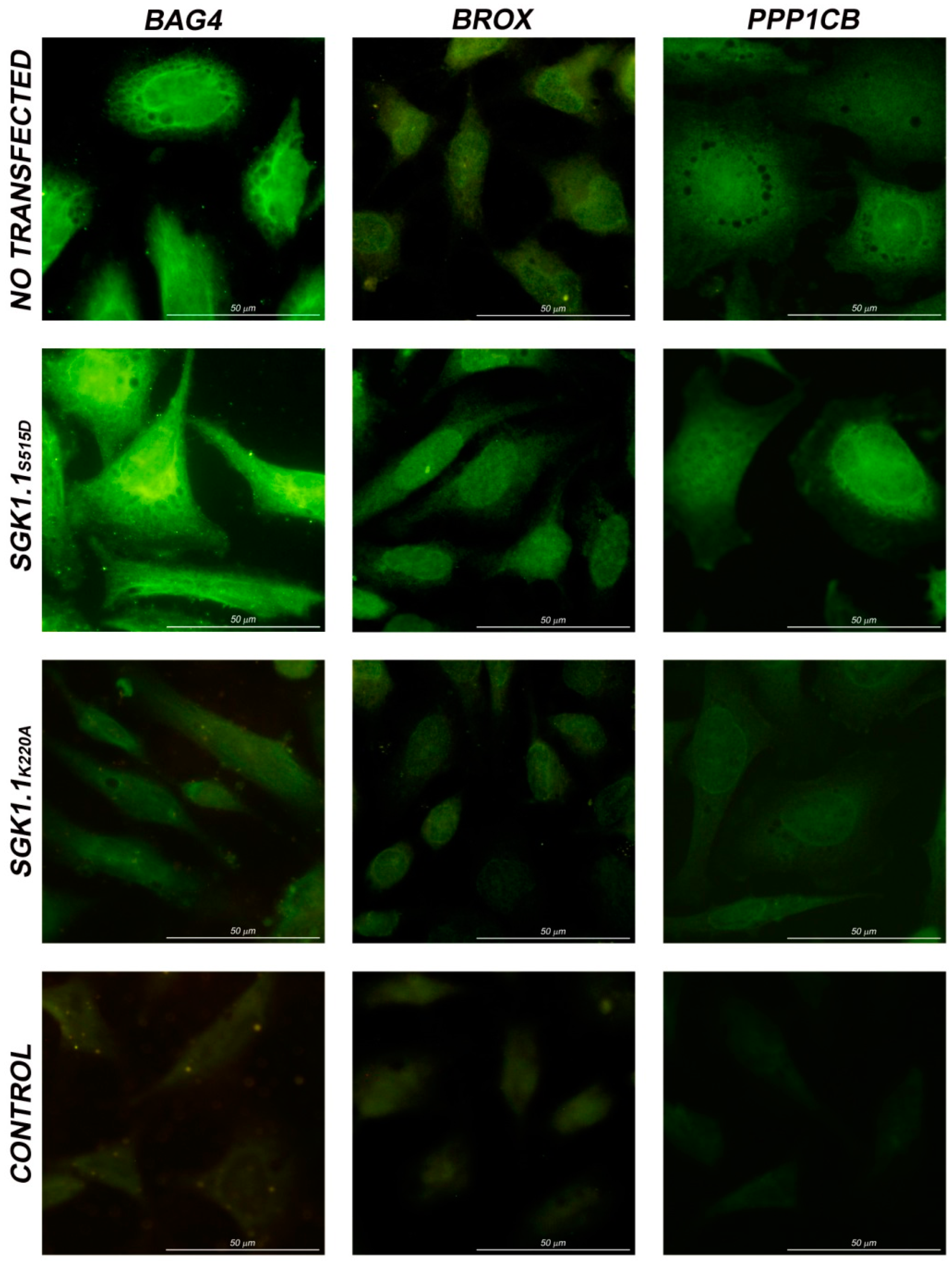
2.4. Immunolocalization in HeLa Cells
3. Experimental Section
3.1. Site Directed Mutagenesis
3.2. Cell Culture and Transfection
3.3. mRNA Extraction from Cells, Double Stranded cDNA Synthesis and PCR Amplification
3.4. ds-cDNA Subtraction and Library Construction
3.5. Dot-Blot Screening
3.6. DNA Sequencing and Sequence Identification
3.7. Gene Expression Analysis by Quantitative RT-PCR (qRT-PCR)
3.8. Plasmid Construction and Preparation of Recombinant Brox Protein
3.9. Production and Purification of Polyclonal Antibodies against the Brox
3.10. Protein Expression Analysis by Western Blot
3.11. Immunofluorescence Microscopic Analyses
3.12. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kobayashi, T.; Deak, M.; Morrice, N.; Cohen, P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem. J. 1999, 344 Pt 1, 189–197. [Google Scholar] [CrossRef]
- Kobayashi, T.; Cohen, P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J. 1999, 339, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.L.R.; Canessa, C.M. Role of SGK in hormonal regulation of epithelial sodium channel in A6 cells. Am. J. Physiol. Cell Physiol. 2003, 284, C404–C414. [Google Scholar] [CrossRef] [PubMed]
- Watt, G.B.; Ismail, N.A.; Caballero, A.G.; Land, S.C.; Wilson, S.M. Epithelial Na+ channel activity in human airway epithelial cells: The role of serum and glucocorticoid-inducible kinase 1. Br. J. Pharmacol. 2012, 166, 1272–1289. [Google Scholar] [CrossRef] [PubMed]
- Baltaev, R.; Strutz-Seebohm, N.; Korniychuk, G.; Myssina, S.; Lang, F.; Seebohm, G. Regulation of cardiac shal-related potassium channel Kv 4.3 by serum- and glucocorticoid-inducible kinase isoforms in Xenopus oocytes. Pflugers Arch. 2005, 450, 26–33. [Google Scholar]
- Boehmer, C.; Laufer, J.; Jeyaraj, S.; Klaus, F.; Lindner, R.; Lang, F.; Palmada, M. Modulation of the voltage-gated potassium channel Kv1.5 by the SGK1 protein kinase involves inhibition of channel ubiquitination. Cell Physiol. Biochem. 2008, 22, 591–600. [Google Scholar]
- Wang, D.; Sun, H.; Lang, F.; Yun, C.C. Activation of NHE3 by dexamethasone requires phosphorylation of NHE3 at Ser663 by SGK1. Am. J. Physiol. Cell Physiol. 2005, 289, C802–C810. [Google Scholar] [CrossRef] [PubMed]
- Loffing, J.; Flores, S.Y.; Staub, O. SGK kinases and their role in epithelial transport. Annu. Rev. Physiol. 2006, 68, 461–490. [Google Scholar] [CrossRef] [PubMed]
- Henke, G.; Maier, G.; Wallisch, S.; Boehmer, C.; Lang, F. Regulation of the voltage gated K+ channel Kv1.3 by the ubiquitin ligase Nedd4–2 and the serum and glucocorticoid inducible kinase SGK1. J. Cell Physiol. 2004, 199, 194–199. [Google Scholar]
- Brunet, A.; Park, J.; Tran, H.; Hu, L.S.; Hemmings, B.A.; Greenberg, M.E. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol. Cell. Biol. 2001, 21, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Luft, F.C. SGK1 survival through various lives may save us all. J. Mol. Med. 2007, 85, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Amato, R.; D’Antona, L.; Porciatti, G.; Agosti, V.; Menniti, M.; Rinaldo, C.; Costa, N.; Bellacchio, E.; Mattarocci, S.; Fuiano, G.; et al. Sgk1 activates MDM2-dependent p53 degradation and affects cell proliferation, survival, and differentiation. J. Mol. Med. 2009, 87, 1221–1239. [Google Scholar]
- You, H.; Jang, Y.; You-Ten, A.I.; Okada, H.; Liepa, J.; Wakeham, A.; Zaugg, K.; Mak, T.W. p53-dependent inhibition of FKHRL1 in response to DNA damage through protein kinase SGK1. Proc. Natl. Acad. Sci. USA 2004, 101, 14057–14062. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Kusakabe, M.; Sunadome, K.; Yamamoto, T.; Nishida, E. The kinase SGK1 in the endoderm and mesoderm promotes ectodermal survival by down-regulating components of the death-inducing signaling complex. Sci. Signal. 2011, 4, ra2. [Google Scholar] [PubMed]
- Zhang, L.; Cui, R.; Cheng, X.; Du, J. Antiapoptotic effect of serum and glucocorticoid-inducible protein kinase is mediated by novel mechanism activating IκB kinase. Cancer Res. 2005, 65, 457–464. [Google Scholar] [PubMed]
- Wang, K.; Gu, S.; Nasir, O.; Foller, M.; Ackermann, T.F.; Klingel, K.; Kandolf, R.; Kuhl, D.; Stournaras, C.; Lang, F. SGK1-dependent intestinal tumor growth in APC-deficient mice. Cell Physiol. Biochem. 2010, 25, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Firestone, G.L.; Giampaolo, J.R.; O’Keeffe, B.A. Stimulus-dependent regulation of serum and glucocorticoid inducible protein kinase (SGK) transcription, subcellular localization and enzymatic activity. Cell Physiol. Biochem. 2003, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lang, F.; Artunc, F.; Vallon, V. The physiological impact of the serum and glucocorticoid-inducible kinase SGK1. Curr. Opin. Nephrol. Hypertens. 2009, 18, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Lang, F.; Cohen, P. Regulation and physiological roles of serum- and glucocorticoid-induced protein kinase isoforms. Sci. STKE 2001, 2001, re17. [Google Scholar] [PubMed]
- Tessier, M.; Woodgett, J.R. Serum and glucocorticoid-regulated protein kinases: Variations on a theme. J. Cell Biochem. 2006, 98, 1391–1407. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, M.F.; Wang, L.; Ravid, T.; Hochstrasser, M.; Canessa, C.M. An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc. Natl. Acad. Sci. USA 2006, 103, 11178–11183. [Google Scholar] [CrossRef] [PubMed]
- Bogusz, A.M.; Brickley, D.R.; Pew, T.; Conzen, S.D. A novel N-terminal hydrophobic motif mediates constitutive degradation of serum- and glucocorticoid-induced kinase-1 by the ubiquitin-proteasome pathway. FEBS J. 2006, 273, 2913–2928. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, M.F.; Coric, T.; Straub, C.; Canessa, C.M. A brain-specific SGK1 splice isoform regulates expression of ASIC1 in neurons. Proc. Natl. Acad. Sci. USA 2008, 105, 4459–4464. [Google Scholar] [CrossRef] [PubMed]
- Raikwar, N.S.; Snyder, P.M.; Thomas, C.P. An evolutionarily conserved N-terminal Sgk1 variant with enhanced stability and improved function. Am. J. Physiol. Renal Physiol. 2008, 295, F1440–F1448. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, M.F.; de la Rosa, A.; Alvarez, J.A.; Canessa, C.M. Multiple translational isoforms give functional specificity to serum- and glucocorticoid-induced kinase 1. Mol. Biol. Cell 2007, 18, 2072–2080. [Google Scholar] [CrossRef] [PubMed]
- Wesch, D.; Miranda, P.; Afonso-Oramas, D.; Althaus, M.; Castro-Hernandez, J.; Dominguez, J.; Morty, R.E.; Clauss, W.; Gonzalez-Hernandez, T.; de la Rosa, A.; et al. The neuronal-specific SGK1.1 kinase regulates δ-epithelial Na+ channel independently of PY motifs and couples it to phospholipase C signaling. Am. J. Physiol Cell Physiol. 2010, 299, C779–C790. [Google Scholar]
- Gey, G.O.; Coffman, W.D.; Kubicek, M.T. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952, 12, 264. [Google Scholar]
- Doong, H.; Vrailas, A.; Kohn, E.C. What’s in the “BAG”?—A functional domain analysis of the BAG-family proteins. Cancer Lett. 2002, 188, 25–32. [Google Scholar]
- Kabbage, M.; Dickman, M.B. The BAG proteins: A ubiquitous family of chaperone regulators. Cell Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Ichioka, F.; Kobayashi, R.; Katoh, K.; Shibata, H.; Maki, M. Brox, a novel farnesylated Bro1 domain-containing protein that associates with charged multivesicular body protein 4 (CHMP4). FEBS J. 2008, 275, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Mu, R.; Dussupt, V.; Jiang, J.; Sette, P.; Rudd, V.; Chuenchor, W.; Bello, N.F.; Bouamr, F.; Xiao, T.S. Two distinct binding modes define the interaction of Brox with the C-terminal tails of CHMP5 and CHMP4B. Structure 2012, 20, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Ceulemans, H.; Bollen, M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004, 84, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Dehner, M.; Hadjihannas, M.; Weiske, J.; Huber, O.; Behrens, J. Wnt signaling inhibits Forkhead box O3a-induced transcription and apoptosis through up-regulation of serum- and glucocorticoid-inducible kinase 1. J. Biol. Chem. 2008, 283, 19201–19210. [Google Scholar] [CrossRef] [PubMed]
- Heikamp, E.B.; Patel, C.H.; Collins, S.; Waickman, A.; Oh, M.H.; Sun, I.H.; Illei, P.; Sharma, A.; Naray-Fejes-Toth, A.; Fejes-Toth, G.; et al. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nat. Immunol. 2014, 15, 457–464. [Google Scholar]
- Burgon, J.; Robertson, A.L.; Sadiku, P.; Wang, X.; Hooper-Greenhill, E.; Prince, L.R.; Walker, P.; Hoggett, E.E.; Ward, J.R.; Farrow, S.N.; et al. Serum and glucocorticoid-regulated kinase 1 regulates neutrophil clearance during inflammation resolution. J. Immunol. 2014, 192, 1796–1805. [Google Scholar]
- Eaton, D.C.; Malik, B.; Bao, H.F.; Yu, L.; Jain, L. Regulation of epithelial sodium channel trafficking by ubiquitination. Proc. Am. Thorac. Soc. 2010, 7, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Diatchenko, L.; Lau, Y.F.; Campbell, A.P.; Chenchik, A.; Moqadam, F.; Huang, B.; Lukyanov, S.; Lukyanov, K.; Gurskaya, N.; Sverdlov, E.D.; et al. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 1996, 93, 6025–6030. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.M.; Crouch, D.H. Cellular distributions of the ERM proteins in MDCK epithelial cells: Regulation by growth and cytoskeletal integrity. Cell Biol. Int. 2001, 25, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Elbaggari, A.; McDonald, K.; Alburo, A. Imaging of chemiluminescent western blots: Comparison of digital imaging and X-ray film. Available online: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_5809.pdf (accessed on 19 March 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Fernández, R.; Ávila, J.; Arteaga, M.F.; Canessa, C.M.; Martín-Vasallo, P. The Neuronal-Specific SGK1.1 (SGK1_v2) Kinase as a Transcriptional Modulator of BAG4, Brox, and PPP1CB Genes Expression. Int. J. Mol. Sci. 2015, 16, 7462-7477. https://doi.org/10.3390/ijms16047462
González-Fernández R, Ávila J, Arteaga MF, Canessa CM, Martín-Vasallo P. The Neuronal-Specific SGK1.1 (SGK1_v2) Kinase as a Transcriptional Modulator of BAG4, Brox, and PPP1CB Genes Expression. International Journal of Molecular Sciences. 2015; 16(4):7462-7477. https://doi.org/10.3390/ijms16047462
Chicago/Turabian StyleGonzález-Fernández, Rebeca, Julio Ávila, María F. Arteaga, Cecilia M. Canessa, and Pablo Martín-Vasallo. 2015. "The Neuronal-Specific SGK1.1 (SGK1_v2) Kinase as a Transcriptional Modulator of BAG4, Brox, and PPP1CB Genes Expression" International Journal of Molecular Sciences 16, no. 4: 7462-7477. https://doi.org/10.3390/ijms16047462







