Interaction of tRNA with Eukaryotic Ribosome
Abstract
:1. Introduction
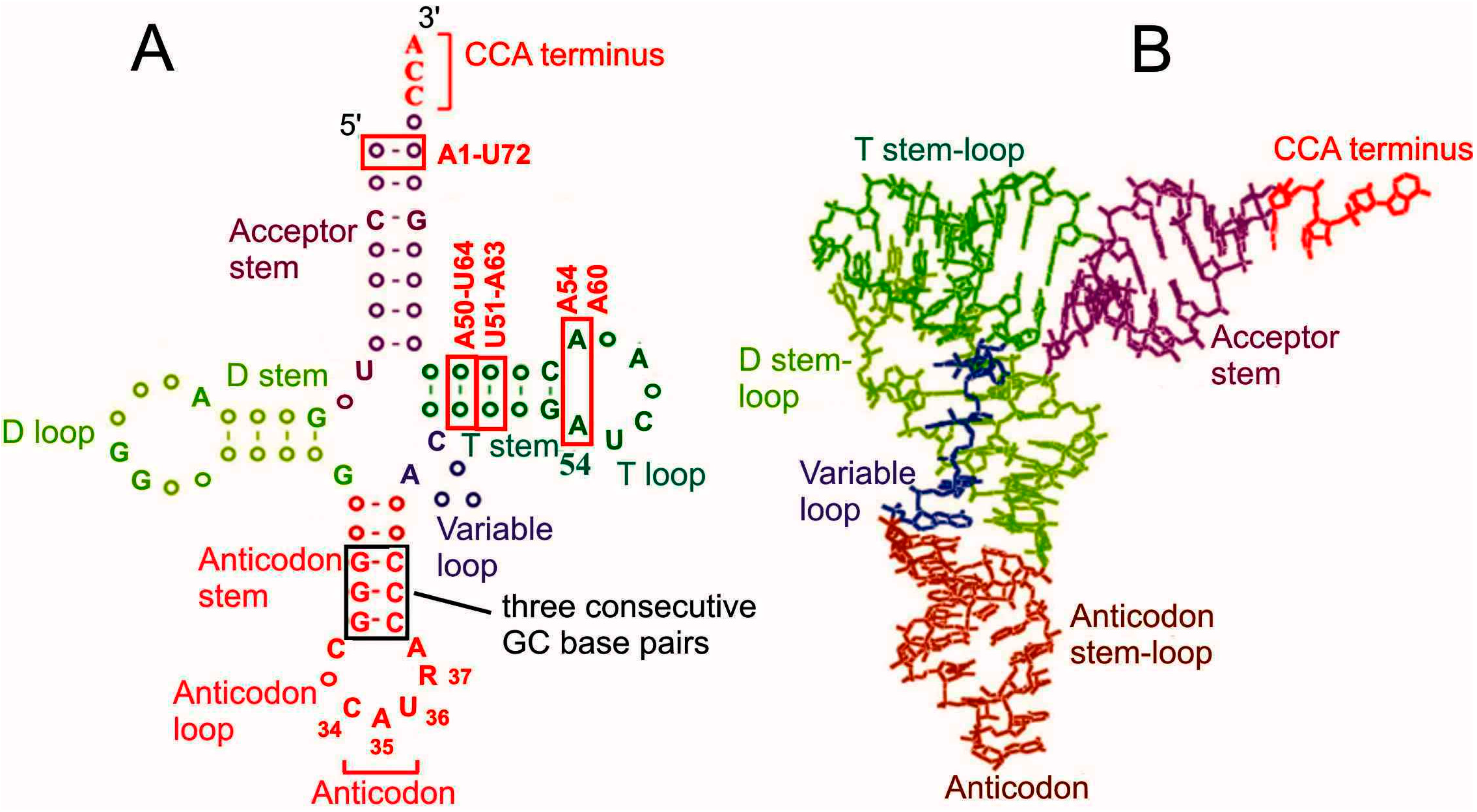
2. The Advance of tRNAs through the A, P and E Sites Produces Hybrid Binding States
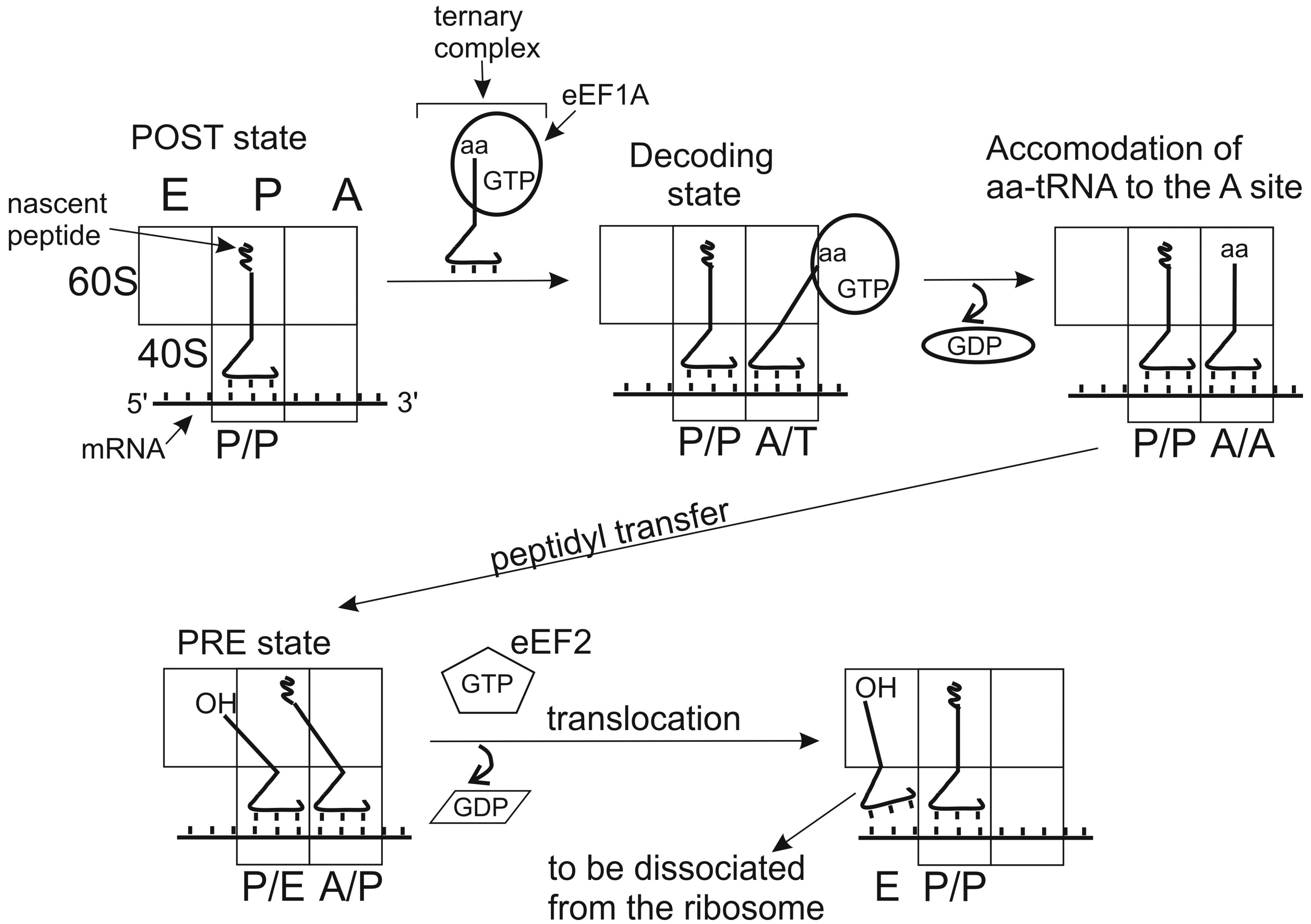
3. Networks of tRNA Interactions Change in the Course of Its Pass through the Stages of Translation Initiation
3.1. Interactions of Met-tRNAi with eIF2
3.2. Interactions of Met-tRNAi in PICs with the 40S Subunit
| tRNA Fragment | Binding Partner | |||
|---|---|---|---|---|
| P Site | E Site | A Site | ||
| Initiation | Elongation | |||
| Anticodon loop | rp uS9 (C-tail) and 18S rRNA h30, h31 and h44 (top) (48S complex [21]) | rp uS9 (C-tail) and 18S rRNA h30, h31 and h44 (top) [14,15,18,20] | 18S rRNA h28, h29, loop h29-h42 and rp uS11 [14] | 18S rRNA 530 loop, h34 and h44 (top) [14] |
| Anticodon stem | 18S rRNA h24, h28 and h29 (48S complex [21]) | 18S rRNA h24a, h30, h43 and h44 (top) [14,15] | 18S rRNA h23b, loop 690 [14] and rp uS5 [14,18,20] | 18S rRNA h18, h30, 965 loop, rp uS19, rp uS12 and/or rp eS30 [14] |
| D stem-loop | N-terminal domain 2 (D2) of eIF2α (43S complex [16]) | 28S rRNA H69 [15,18,20] and rp eL44 (C-tail) [18,20] | 28S rRNA loop H77-H78 ** [14] | 28S rRNA H69, H38 (“A site finger”) [14], rp uS12 and H89 in A/T state [20] |
| T stem-loop | rp uL5 * [15,18,20], eL44 (C-tail) [18,20] and 28S rRNA H85, loop H82-H83 * [14] | rp uL1 ** [14,18,20] and 28S rRNA loop H77-H78 ** [14] | 28S rRNA H89 [14], rp uS12 and sarcin-ricin loop (apical loop of 28S rRNA H95) in A/T state [20] | |
| Acceptor stem | C-terminal domain of eIF5B [6,19] | 28S rRNA H80 * [14,15] and H93 [15] | 28S rRNA H68 ** [14] | 28S rRNA loop H69-H71 [14] |
| CCA-end | 28S rRNA H80 [6], and domain IV of eIF5B [6,19] (80S complex) | 28S rRNA H93, the PTC ring [14,25] and H89 [25] | 28S rRNA H82 [18], loop H74-H88 ** [25] and rp eL44 ** (region of K53 in human numbering) [18,20,23,24] | 28S rRNA H89 [14,25], H92, and H93 [25] |
| 5'-end | not analyzed | Rp uL16 [15,20] | not analyzed | not analyzed |
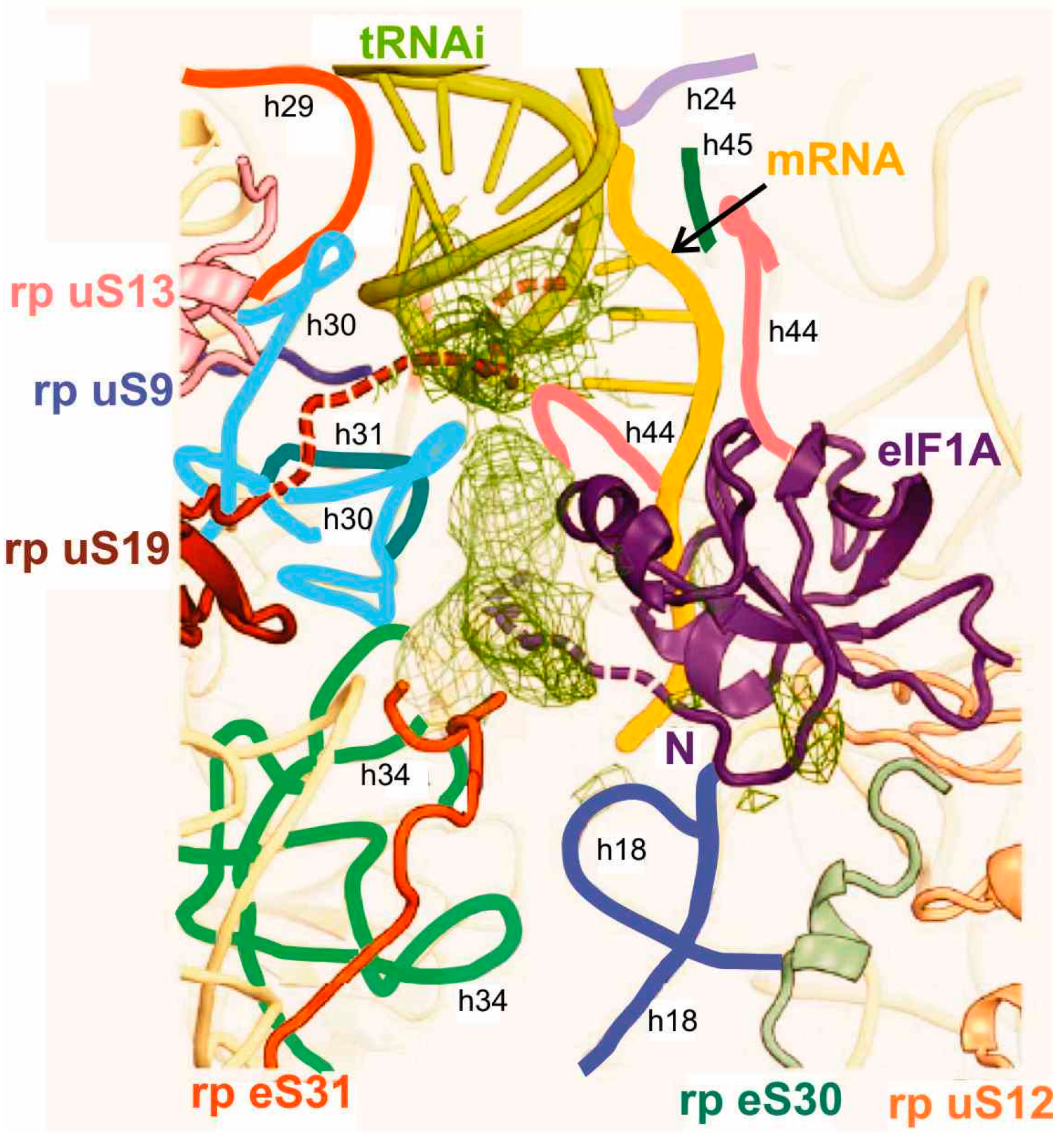
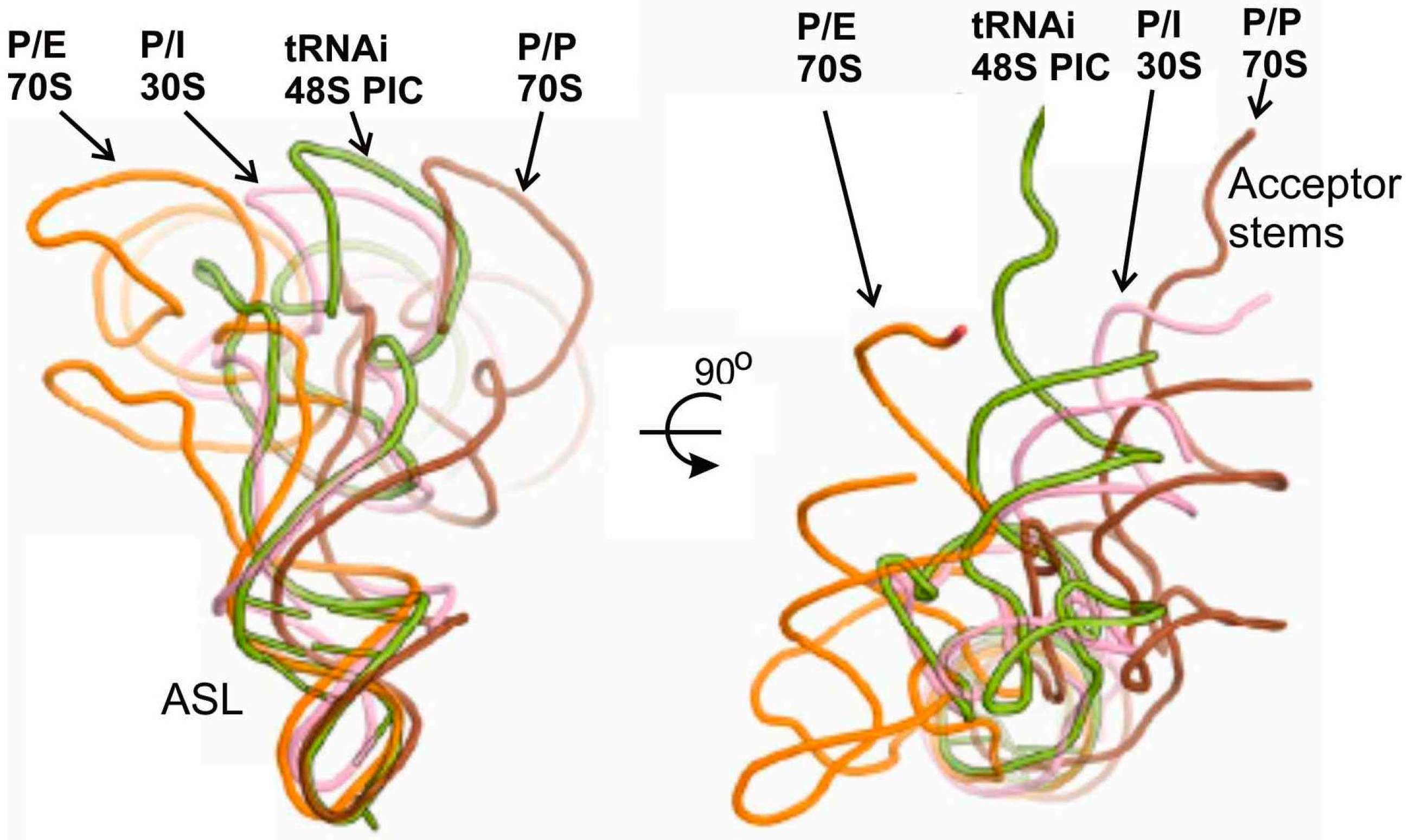
3.3. Interactions of Met-tRNAi in 80S Initiation Complexes
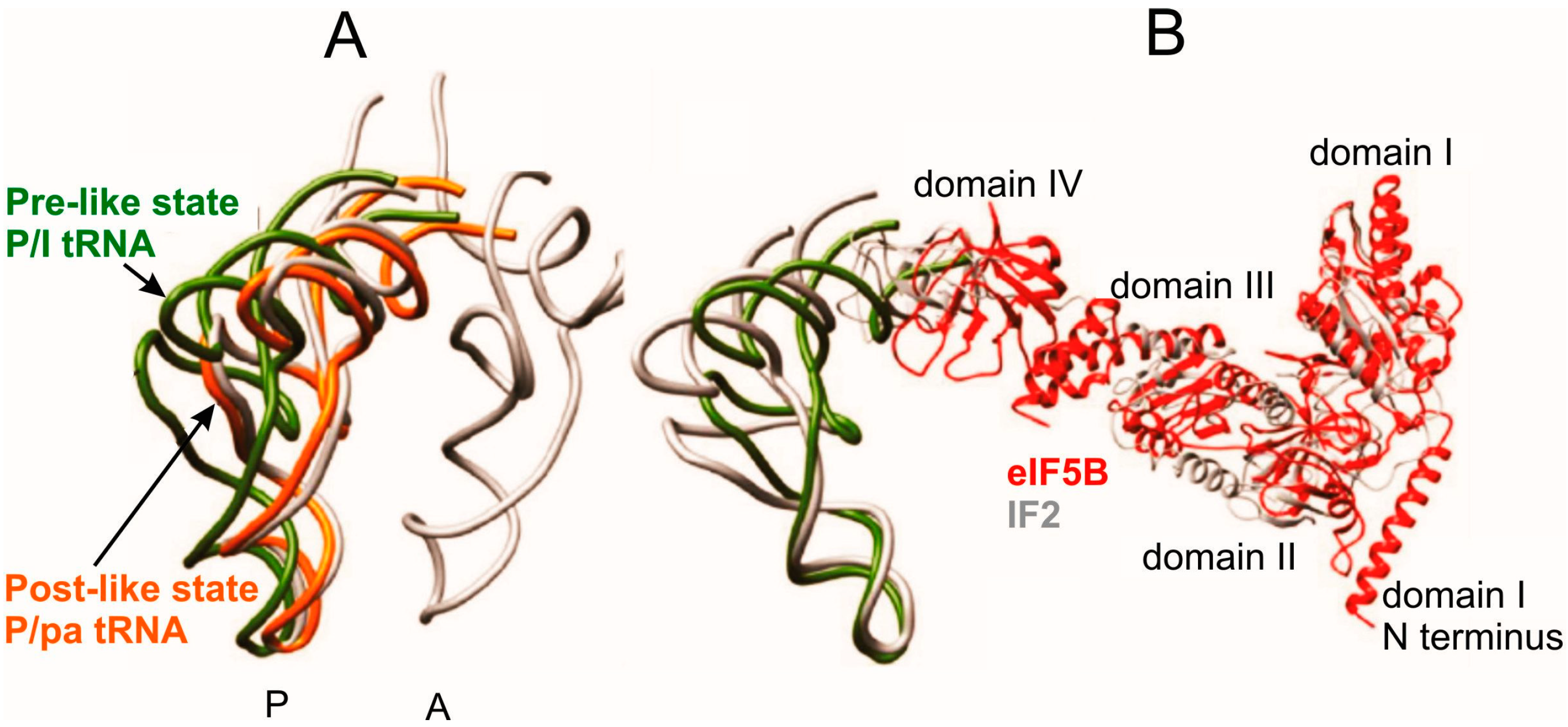
4. Dynamics of tRNA Interactions during Translation Elongation
4.1. Interactions in POST Complexes
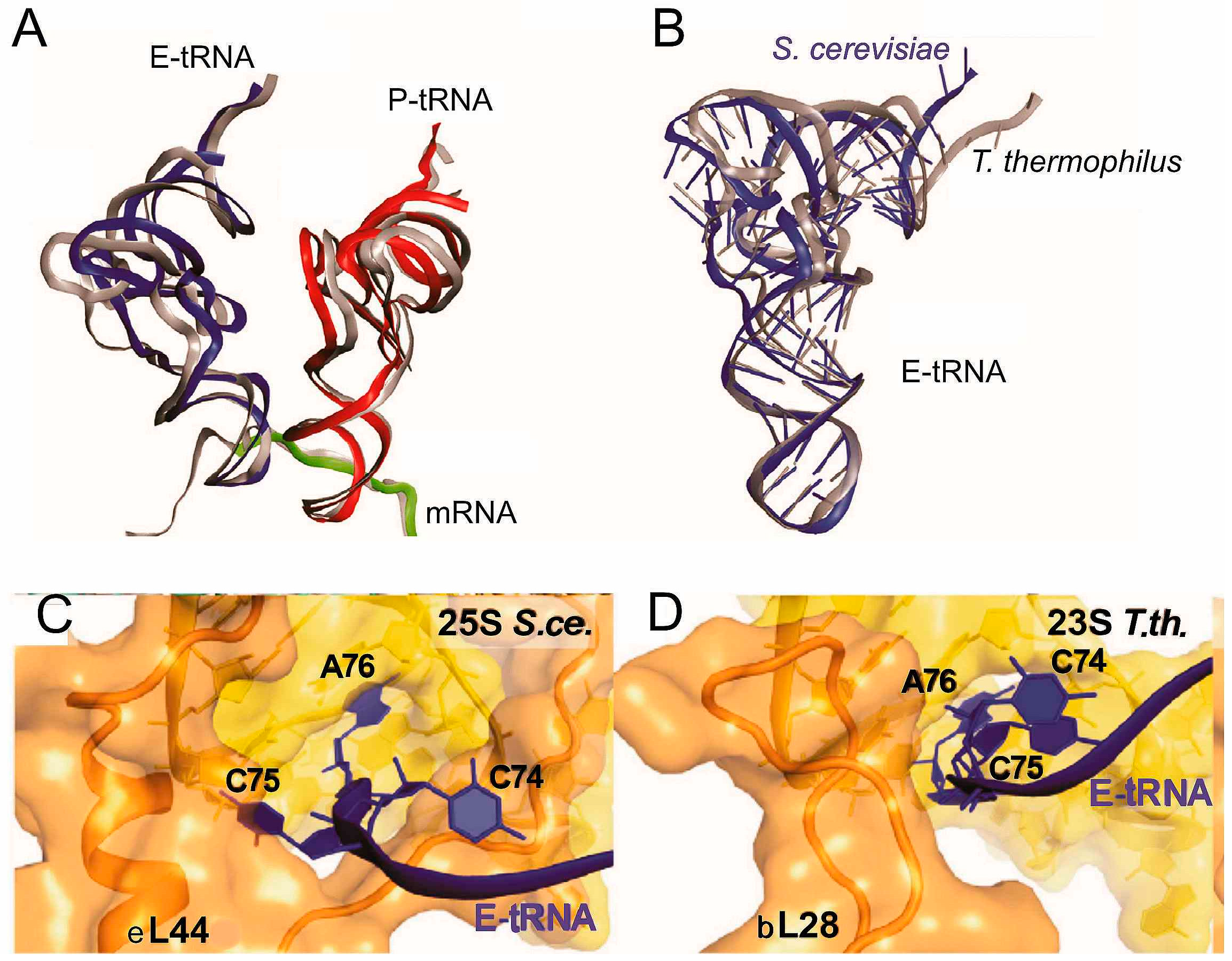
4.2. Interactions in PRE Complexes
4.3. Interactions in Model 80S Complexes with a Single Deacylated tRNA at the P Site
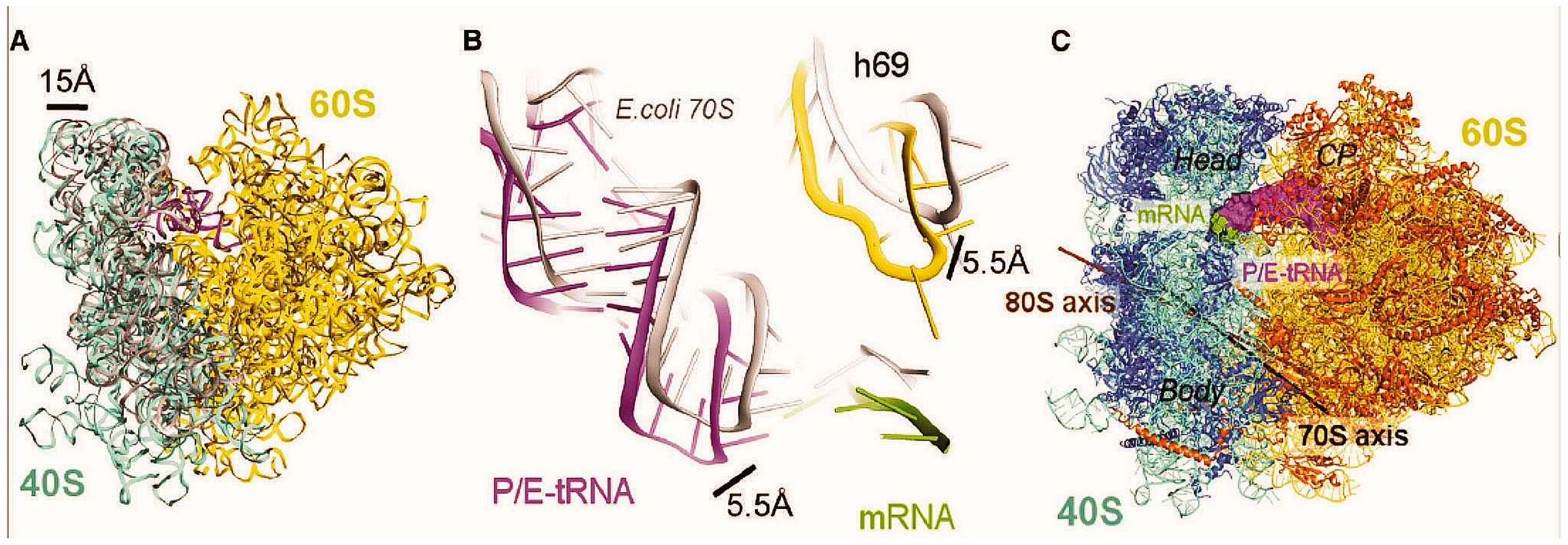
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Watson, J.D.; Baker, T.A.; Bell, S.P.; Gann, A.; Levine, M.; Oosick, R. Molecular Biology of the Gene; Pearson PLC: San Francisco, CA, USA, 2008. [Google Scholar]
- Rasmussen, L.C.; Laursen, B.S.; Mortensen, K.K.; Sperling-Petersen, H.U. Initiator tRNAs in bacteria and eukaryotes. eLS 2009. [Google Scholar] [CrossRef]
- Kolitz, S.E.; Lorsch, J.R. Eukaryotic initiator tRNA: Finely tuned and ready for action. FEBS Lett. 2010, 584, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Jühling, F.; Püetz, J.; Stadler, P.; Sauter, C.; Florentz, C. Structure of transfer RNAs: Similarity and variability. WIREs RNA 2012, 3, 37–61. [Google Scholar] [CrossRef] [PubMed]
- Hori, H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 2014, 5, 144. [Google Scholar] [CrossRef]
- Fernandez, I.S.; Bai, X.-C.; Hussain, T.; Kelley, A.C.; Lorsch, J.R.; Ramakrishnan, V.; Scheres, S.H.W. Molecular architecture of a eukaryotic translational initiation complex. Science 2013, 342, 1240585. [Google Scholar] [CrossRef] [PubMed]
- Kapp, L.D.; Lorsch, J.R. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 2004, 335, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Hinnebush, A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011, 75, 434–467. [Google Scholar] [CrossRef] [PubMed]
- Transfer RNA database. Available online: http://trna.bioinf.uni-leipzig.de/ (accessed on 21 February 2015).
- Rodnina, M.V.; El’skaya, A.V.; Semenkov, Y.P.; Kirillov, S.V. Number of tRNA binding sites on 80 S ribosomes and their subunits. FEBS Lett. 1988, 231, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Rodnina, M.V.; El’skaya, A.V.; Semenkov, Y.P.; Kirillov, S.V. Interaction of tRNA with the A and P sites of rabbit-liver 80s ribosomes and their 40S subunits. Eur. J. Biochem. 1989, 185, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Graifer, D.M.; Nekhai, S.Y.; Mundus, D.A.; Fedorova, O.S.; Karpova, G.G. Interaction of human and Escherichia coli tRNA with human 80S ribosomes in the presence of oligo- and polyuridylate template. Biochim. Biophys. Acta 1992, 1171, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Budkevich, T.V.; El’skaya, A.V.; Nierhaus, K.H. Features of 80S mammalian ribosome and its subunits. Nucleic Acids Res. 2008, 36, 4736–4744. [Google Scholar] [CrossRef] [PubMed]
- Budkevich, T.; Giesebrecht, J.; Altman, R.B.; Munro, J.B.; Mielke, T.; Nierhaus, K.H.; Blanchard, S.C.; Spahn, C.M. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell 2011, 44, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Spahn, C.M.; Beckmann, R.; Eswar, N.; Penczek, P.A.; Sali, A.; Blobel, G.; Frank, J. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell 2001, 107, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Hashem, Y.; desGeorges, A.; Dhote, V.; Langlois, R.; Liao, H.Y.; Grassucci, R.A.; Hellen, C.U.T.; Pestova, T.V.; Frank, J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 2013, 153, 1108–1119. [Google Scholar] [CrossRef] [PubMed]
- Anger, A.M.; Armache, J.-P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 49, 80–87. [Google Scholar] [CrossRef]
- Svidritskiy, E.; Brilot, A.F.; Koh, C.S.; Grigorieff, N.; Korostelev, A.A. Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations. Structure 2014, 22, 1210–1218. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Unbehaun, A.; Loerke, J.; Behrmann, E.; Collier, M.; Burger, J.; Mielke, T.; Spahn, C.M.T. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 2014, 21, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Budkevich, T.V.; Giesebrecht, J.; Behrmann, E.; Loerke, J.; Ramrath, D.J.F.; Mielke, T.; Ismer, J.; Hildebrand, P.W.; Tung, C-S.; Nierhaus, K.H.; et al. Regulation of the mammalian elongation cycle by subunit rolling: A eukaryotic-specific ribosome rearrangement. Cell 2014, 158, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Lomakin, I.B.; Steitz, T.A. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 2013, 500, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Bulygin, K.; Favre, A.; Baouz-Drahy, S.; Hountondji, C.; Vorobjev, Y.; Ven’yaminova, A.; Graifer, D.; Karpova, G. Arrangement of 3'-terminus of tRNA on the human ribosome as revealed from cross linking data. Biochimie 2008, 90, 1624–1636. [Google Scholar] [CrossRef] [PubMed]
- Baouz, S.; Woisard, A.; Sinapah, S.; Le Caer, J.P.; Argentini, M.; Bulygin, K.; Aguie, G.; Hountondji, C. The human large subunit ribosomal protein L36A-like contacts the CCA end of P-site bound tRNA. Biochimie 2009, 91, 1420–1425. [Google Scholar] [CrossRef] [PubMed]
- Hountondji, C.; Bulygin, K.; Woisard, A.; Tuffery, P.; Crechet, J.-B.; Pech, M.; Nierhaus, K.H.; Karpova, G.; Baouz, S. Lys53 of ribosomal protein L36AL and the CCA end of a tRNA at the P/E hybrid Site are in close proximity on the human ribosome. Chembiochem 2012, 13, 1791–1797. [Google Scholar] [CrossRef] [PubMed]
- Bulygin, K.; Malygin, A.; Hountondji, C.; Graifer, D.; Karpova, G. Positioning of CCA-arms of the A- and the P-tRNAs towards the 28S rRNA in the human ribosome. Biochimie 2013, 95, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Beckmann, R.; Cate, J.H.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014, 24, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Kirillov, S.V.; Kemkhadze, K.S.; Makarov, E.M.; Makhno, V.I.; Odintsov, V.B.; Semenkov, Y.P. Mechanism of codon-anticodon interaction in ribosomes: Codon-anticodon interaction of aminoacyl-tRNA at the ribosomal donor site. FEBS Lett. 1980, 120, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Kirillov, S.V.; Semenkov, Y.P. Non-exclusion principle of Ac-Phe-tRNAPhe interaction with the donor and acceptor sites of Escherichia coli ribosomes. FEBS Lett. 1982, 148, 235–238. [Google Scholar] [CrossRef]
- Kirillov, S.V.; Makarov, E.M.; Semenkov, Y.P. Quantitative study of interaction of deacylated tRNA with Escherichia coli ribosomes. Role of 50 S subunits in formation of the E site. FEBS Lett. 1983, 157, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Gnirke, A.; Nierhaus, K.H. tRNA binding sites on the subunits of Escherichia coli ribosomes. J. Biol. Chem. 1986, 261, 14506–14514. [Google Scholar] [PubMed]
- Rheinberger, H.J.; Nierhaus, K.H. Adjacent codon-anticodon interactions of both tRNAs present at the ribosomal A and P or P and E sites. FEBS Lett. 1986, 204, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Schilling-Bartetzko, S.; Bartetzko, A.; Nierhaus, K.H. Kinetic and thermodynamic parameters for tRNA binding to the ribosome and for the translocation reaction. J. Biol. Chem. 1992, 267, 4703–4712. [Google Scholar] [PubMed]
- Demeshkina, N.; Repkova, M.; Ven’yaminova, A.; Graifer, D.; Karpova, G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P and E sites, respectively: A cross-linking study with mRNA analogues carrying aryl azide group at either the uracil or the guanine residue. RNA 2000, 6, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Graifer, D.; Molotkov, M.; Styazhkina, V.; Demeshkina, N.; Bulygin, K.; Eremina, A.; Ivanov, A.; Laletina, E.; Ven’yaminova, A.; Karpova, G. Variable and conserved elements of human ribosomes surrounding the mRNA at the decoding and upstream sites. Nucleic Acids Res. 2004, 32, 3282–3293. [Google Scholar] [PubMed]
- Yusupov, M.M.; Yusupova, G.Zh.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.D.H.; Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Yusupova, G.Z.; Yusupov, M.M.; Cate, J.H.D.; Noller, H.F. The path of messenger RNA through the ribosome. Cell 2001, 106, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Selmer, M.; Dunham, C.M.; Murphy, F.V., 4th; Weixlbaumer, A.; Petry, S.; Kelley, A.C.; Weir, J.R.; Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313, 1935–1942. [Google Scholar] [CrossRef] [PubMed]
- Jenner, L.B.; Demeshkina, N.; Yusupova, G.; Yusupov, M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010, 17, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, S.C.; Kim, H.D.; Gonzalez, R.L., Jr.; Puglisi, J.D.; Chu, S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA 2004, 101, 12893–12898. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.D.; Puglisi, J.D.; Chu, S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys. J. 2007, 93, 3575–3582. [Google Scholar] [CrossRef] [PubMed]
- Munro, J.B.; Altman, R.B.; O’Connor, N.; Blanchard, S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell 2007, 25, 505–517. [Google Scholar] [PubMed]
- Moazed, D.; Noller, H.F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 1989, 57, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Moazed, D.; Noller, H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature 1989, 342, 142–148. [Google Scholar] [CrossRef]
- Caulfield, T.; Devkota, B. Motion of transfer RNA from the A/T state into the A-site using docking and simulations. Proteins 2012, 80, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, A.; Marzi, S.; Myasnikov, A.G.; Fabbretti, A.; Yusupov, M.; Gualerzi, C.O.; Klaholz, B.P. Structure of the 30S translation initiation complex. Nature 2008, 455, 416–420. [Google Scholar] [CrossRef]
- Myasnikov, A.G.; Simonetti, A.; Marzi, S.; Klaholz, B.P. Structure–function insights into prokaryotic and eukaryotic translation initiation. Curr. Opin. Struct. Biol. 2009, 19, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N.; Doudna Cate, J.H. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef]
- Ratje, A.H.; Loerke, J.; Mikolajka, A.; Brueunner, M.; Hildebrand, P.W.; Starosta, A.L.; Doenhoefer, A.; Connell, S.R.; Fucini, P.; Mielke, T.; et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 2010, 468, 713–716. [Google Scholar] [CrossRef] [PubMed]
- Dunkle, J.A.; Wang, L.; Feldman, M.B.; Pulk, A.; Chen, V.B.; Kapral, G.J.; Noeske, J.; Richardson, J.S.; Blanchard, S.C.; Cate, J.H. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 2011, 332, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Rodnina, M.; Wintermeyer, W. Recent mechanistic insights into eukaryotic ribosomes. Curr. Opin. Cell Biol. 2009, 21, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, E.; Panvert, M.; Lazennec-Schurdevin, C.; Coureux, P.D.; Perez, J.; Thompson, A.; Mechulam, Y. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat. Struct. Mol. Biol. 2012, 19, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Stolboushkina, E.; Nikonov, S.; Zelinskaya, N.; Arkhipova, V.; Nikulin, A.; Garber, M.; Nikonov, O. Crystal structure of the archaeal translation initiation factor 2 in complex with a GTP analogue and Met-tRNAfMet. J. Mol. Biol. 2013, 425, 989–998. [Google Scholar] [CrossRef]
- Yatime, L.; Schmitt, E.; Blanquet, S.; Mechulam, Y. Functional molecular mapping of archaeal translation initiation factor 2. J. Biol. Chem. 2004, 279, 15984–15993. [Google Scholar] [CrossRef] [PubMed]
- Pedulla, N.; Palermo, R.; Hasenohrl, D.; Blasi, U.; Cammarano, P.; Londei, P. The archaeal eIF2 homologue: Functional properties of an ancient translation initiation factor. Nucleic Acids Res. 2005, 33, 1804–1812. [Google Scholar] [CrossRef] [PubMed]
- Yatime, L.; Mechulam, Y.; Blanquet, S.; Schmitt, E. Structural switch of the gamma subunit in an archaeal aIF2 alpha gamma heterodimer. Structure 2006, 14, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Naveau, M.; Lazennec-Schurdevin, C.; Panvert, M.; Dubiez, E.; Mechulam, Y.; Schmitt, E. Roles of yeast eIF2α and eIF2β subunits in the binding of the initiator methionyl-tRNA. Nucleic Acids Res. 2013, 41, 1047–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharifulin, D.; Babaylova, E.; Kossinova, O.; Bartuli, Y.; Graifer, D.; Karpova, G. Ribosomal protein S5e is implicated in translation initiation via its interaction with N-terminal domain of initiation factor eIF2α. Chembiochem 2013, 14, 2136–2143. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Llacer, J.L.; Fernaґndez, I.S.; Munoz, A.; Martin-Marcos, P.; Savva, C.G.; Lorsch, J.R.; Hinnebusch, A.G.; Ramakrishnan, V. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 2014, 159, 597–607. [Google Scholar] [CrossRef]
- Saini, A.K.; Nanda, J.S.; Lorsch, J.R.; Hinnebusch, A.G. Regulatory elements in eIF1A control the fielity of start codon selection by modulating tRNA(i) (Met) binding to the ribosome. Genes Dev. 2010, 24, 97–110. [Google Scholar] [CrossRef]
- Allen, G.S.; Zavialov, A.; Gursky, R.; Ehrenberg, M.; Frank, J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 2005, 121, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Julian, P.; Milon, P.; Agirrezabala, X.; Lasso, G.; Gil, D.; Rodnina, M.V.; Valle, M. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011, 9, e1001095. [Google Scholar] [CrossRef]
- Noller, H.F.; Hoang, L.; Fredrick, K. The 30S ribosomal P site: A function of 16S rRNA. FEBS Lett. 2005, 579, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Cate, J.H.; Yusupov, M.M.; Yusupova, G.Z.; Earnest, T.N.; Noller, H.F. X-ray crystal structures of 70S ribosome functional complexes. Science 1999, 285, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Korostelev, A.; Trakhanov, S.; Laurberg, M.; Noller, H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 2006, 126, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.; Moore, P.B.; Steitz, T.A. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 2003, 9, 1345–1352. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Weixlbaumer, A.; Loakes, D.; Kelley, A.C.; Ramakrishnan, V. Insights into substrate stabilization from snapshots of the peptidyltransferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009, 16, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Agirrezabala, X.; Lei, J.; Brunelle, J.L.; Ortiz-Meoz, R.F.; Green, R.; Frank, J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol. Cell 2008, 32, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Julian, P.; Konevega, A.L.; Scheres, S.H.; Lazaro, M.; Gil, D.; Wintermeyer, W.; Rodnina, M.V.; Valle, M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc. Natl. Acad. Sci. USA 2008, 105, 16924–16927. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Schmeing, T.M.; Kelley, A.C.; Ramakrishnan, V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 2010, 330, 835–838. [Google Scholar] [PubMed]
- Ogle, J.M.; Murphy, F.V.; Tarry, M.J.; Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 2002, 111, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Voorhees, R.M.; Kelley, A.C.; Gao, Y.G.; Murphy, F.V., 4th; Weir, J.R.; Ramakrishnan, V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 2009, 326, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Schuette, J.C.; Murphy, F.V., 4th; Kelley, A.C.; Weir, J.R.; Giesebrecht, J.; Connell, S.R.; Loerke, J.; Mielke, T.; Zhang, W.; Penczek, P.A.; et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009, 28, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Munro, J.B.; Sanbonmatsu, K.Y.; Spahn, C.M.; Blanchard, S.C. Navigating the ribosome’s metastable energy landscape. Trends Biochem. Sci. 2009, 34, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Graifer, D.; Karpova, G. Photoactivatable RNA derivatives as tools for studying the structural and functional organization of complex cellular ribonucleoprotein machineries. RSC Adv. 2013, 3, 2858–2872. [Google Scholar] [CrossRef]
- Graifer, D.; Zhigailov, A.; Ven'yaminova, A.; Malygin, A.; Iskakov, B.; Karpova, G. 2'-OH of mRNA are critical for the binding of its codons at the 40S ribosomal P site but not at the mRNA entry site. FEBS Lett. 2012, 586, 3731–3736. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graifer, D.; Karpova, G. Interaction of tRNA with Eukaryotic Ribosome. Int. J. Mol. Sci. 2015, 16, 7173-7194. https://doi.org/10.3390/ijms16047173
Graifer D, Karpova G. Interaction of tRNA with Eukaryotic Ribosome. International Journal of Molecular Sciences. 2015; 16(4):7173-7194. https://doi.org/10.3390/ijms16047173
Chicago/Turabian StyleGraifer, Dmitri, and Galina Karpova. 2015. "Interaction of tRNA with Eukaryotic Ribosome" International Journal of Molecular Sciences 16, no. 4: 7173-7194. https://doi.org/10.3390/ijms16047173





