Fatal Attraction: How Bacterial Adhesins Affect Host Signaling and What We Can Learn from Them
Abstract
:1. Introduction
2. Bacterial Adhesin Classes and Their Ligands
2.1. Integrin and Fibronectin Binding Proteins
| Organism | Adhesin | Ligand | Function | Refs. |
|---|---|---|---|---|
| S. aureus | Clumping factor A (ClfA) | Fibrinogen γ-chain | Adhesion and immune evasion | [14,15] |
| Clumping factor B (ClfB) | Fibrinogen α-chain, keratin 10 and loricrin | Adhesion to desquamated epithelial cells | [15] | |
| FnBPA/FnBPB | Fibronectin, Fibrinogen γ-chain and elastin | Adhesion to ECM, biofilm formation | [15] | |
| Collagen adhesin (Cna) | Collagen, complement C1q | Adhesion, complement evasion | [15] | |
| Streptococcal sp. | Sfbl | Fibronectin | Adhesion | [8,9] |
| Yersinia sp. | Invasin | β1-integrin | Adhesion, internalization | [16] |
| Trimeric autotransporter YadA | Fibronectin, Collagen | Adhesion, internalization | [17] | |
| Ail | Fibronectin, Laminin, C4bp, complement H | Yop delivery, adhesion, internalization, serum resistance | [18,19] | |
| E. coli | CU P-pilus | Gal(α1-4)gal containing receptors | Adhesion, immune response | [20] |
| CU type I pili | Mannose containing glycoproteins | Adhesion, inflammation | [21,22] | |
| Afa/Dr | Collagen, hDAF, CEACAMs | Adhesion, inflammation | [23] | |
| Curli | Fibronectin, laminin | Biofilm formation, invasion, inflammation | [24] | |
| Trimeric autotransporter Antigen 43 | Unknown | Aggregation | [25] | |
| N. meningitidis | Type IV pilus | Unknown | Adhesion, aggregation, motility, DNA transfer | [26] |
| M. tuberculosis | Mtp amyloid | Laminin | Adhesion, colonization | [27] |
| MCE1a | Unknown | Adhesion, invasion | [28,29] | |
| V. parahaemolyticus | MAM7 | Phosphatidic acid, fibronectin | Adhesion, invasion | [30] |
| H. pylori | Type IV pilus | β5-Integrin | Gastrin production, increases acidity | [31] |
| BabA | Lewis B antigen | Adhesion, inflammation | [32] | |
| L. rhamnosus GG | SpaCBA pilus | Mucus | Adhesion, immunomdulation | [33,34] |
| Salmonella sp. | FliC | Cholesterol | Adhesion, biofilm formation | [35] |
| PefA | Lewis X blood group antigen | Adhesion | [36] | |
| Type I pilus FimH | Mannose containing glycoproteins | Adhesion | [37] |
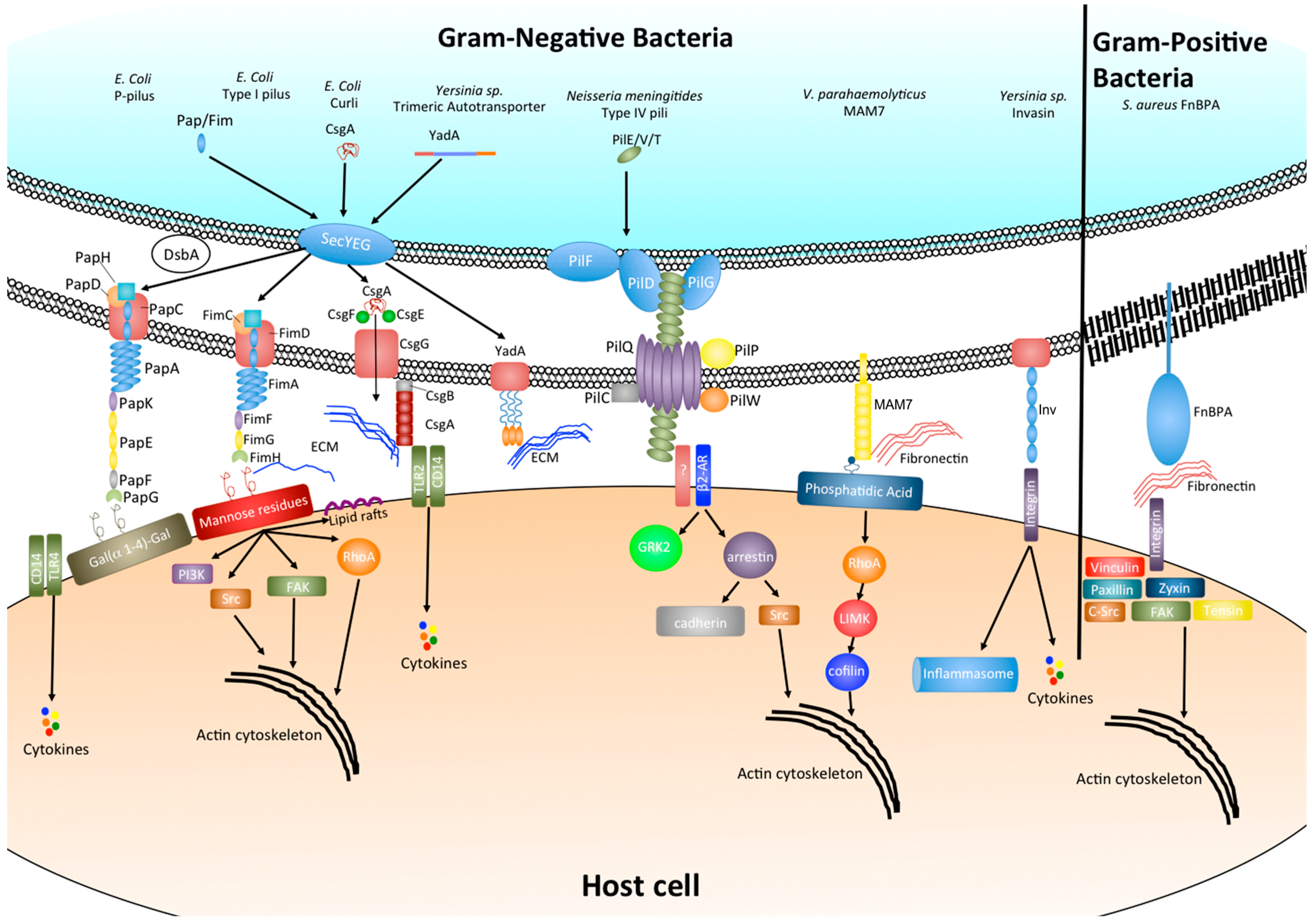
2.2. Chaperone-Usher Pili: P Pili and Type I Pili
2.3. Type IV Pili
2.4. Adhesive Amyloids
2.5. Autotransporters
2.6. Multivalent Adhesion Molecules
3. Effect of Bacterial Adhesion on Host Cell Signaling
4. The Potential of Adhesion Inhibition as Novel Infection Intervention
5. Summary
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pizarro-Cerdá, J.; Cossart, P. Bacterial adhesion and entry into host cells. Cell 2006, 124, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.A.; Fälker, S.; Dahlberg, S.; Normark, S.; Henriques-Normark, B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 2009, 5, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Ohlsen, K.; Hauck, C.R. Intergrin-mediated uptake of fibronectin-binding bacteria. Eur. J. Cell Biol. 2011, 90, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Simakov, O.; Chapman, J.; Fahey, B.; Gauthier, M.E.A.; Mitros, T.; Richards, G.S.; Conaco, C.; Dacre, M.; Hellsten, U.; et al. The amphimedon queenslandica genome and the evolution of animal complexity. Nature 2010, 466, 720–726. [Google Scholar] [CrossRef]
- Patti, J.M.; Allen, B.L.; McGavin, M.J.; Hook, M. Mscramm-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 1994, 48, 585–617. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.; François, P.P.; Nüße, O.; Foti, M.; Hartford, O.M.; Vaudaux, P.; Foster, T.J.; Lew, D.P.; Herrmann, M.; Krause, K.H. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell. Microbiol. 1999, 1, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Molinari, G.; Talay, S.R.; Valentin-Weigand, P.; Rohde, M.; Chhatwal, G.S. The fibronectin-binding protein of streptococcus pyogenes, Sfbi, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun. 1997, 65, 1357–1363. [Google Scholar] [PubMed]
- Ozeri, V.; Rosenshine, I.; Mosher, D.F.; Fässler, R.; Hanski, E. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol. Microbiol. 1998, 30, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Herman, P.; El-Kirat-Chatel, S.; Beaussart, A.; Geoghegan, J.A.; Foster, T.J.; Dufrene, Y.F. The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol. Microbiol. 2014, 93, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Bingham, R.J.; Rudiño-Piñera, E.; Meenan, N.A.G.; Schwarz-Linek, U.; Turkenburg, J.P.; Höök, M.; Garman, E.F.; Potts, J.R. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc. Natl. Acad. Sci. USA 2008, 105, 12254–12258. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Linek, U.; Höök, M.; Potts, J.R. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 2004, 52, 631–641. [Google Scholar] [CrossRef] [PubMed]
- McCourt, J.; O’Halloran, D.P.; McCarthy, H.; O’Gara, J.P.; Geoghegan, J.A. Fibronectin-binding proteins are required for biofilm formation by community-associated methacillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol. Lett. 2014, 353, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, V.K.; Rivera, J.J.; Smeds, E.; Ko, Y.P.; Bowden, M.G.; Wann, E.R.; Gurusiddappa, S.; Fitzgerald, J.R.; Höök, M. A structural model of the Staphylococcus aureus ClfA–fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 2008, 4, e1000226. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Hook, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Isberg, R.R.; Leong, J.M. Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 1990, 60, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Heise, T.; Dersch, P. Identification of a domain in yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc. Natl. Acad. Sci. USA 2006, 103, 3375–3380. [Google Scholar] [CrossRef] [PubMed]
- Tsang, T.M.; Wiese, J.S.; Felek, S.; Kronshage, M.; Krukonis, E.S. Ail proteins of Yersinia pestis and Y. pseudotuberculosis have different cell binding and invasion activities. PLoS One 2013, 8, e83621. [Google Scholar] [CrossRef] [PubMed]
- Felek, S.; Krukonis, E.S. The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect. Immun. 2009, 77, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Wurpel, D.J.; Beatson, S.A.; Totsika, M.; Petty, N.K.; Schembri, M.A. Chaperone-usher fimbriae of Escherichia coli. PLoS One 2013, 8, e52835. [Google Scholar] [CrossRef] [PubMed]
- Lillington, J.; Geibel, S.; Waksman, G. Reprint of “biogenesis and adhesion of type 1 and P-pili”. Biochim. Biophys. Acta 2014, 1850, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Dreux, N.; Denizot, J.; Martinez-Medina, M.; Mellmann, A.; Billig, M.; Kisiela, D.; Chattopadhyay, S.; Sokurenko, E.; Neut, C.; Gower-Rousseau, C.; et al. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 2013, 9, e1003141. [Google Scholar] [CrossRef]
- Servin, A.L. Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): Current insights and future challenges. Clin. Microbiol. Rev. 2014, 27, 823–869. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.L.L.; Kwan, A.H.; Sunde, M. Functional amyloid: Widespread in nature, diverse in purpose. Essays Biochem. 2014, 56, 207–209. [Google Scholar] [CrossRef] [PubMed]
- Heras, B.; Totsika, M.; Peters, K.M.; Paxman, J.J.; Gee, C.L.; Jarrott, R.J.; Perugini, M.A.; Whitten, A.E.; Schembri, M.A. The Antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc. Natl. Acad. Sci. USA 2014, 111, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Giltner, C.L.; Nguyen, Y.; Burrows, L.L. Type IV pilin proteins: Versatile molecular modules. Microbiol. Mol. Biol. Rev. 2012, 76, 740–772. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Xicohténcatl-Cortes, J.; Hess, S.; Caballero-Olín, G.; Girón, J.A.; Friedman, R.L. Mycobacterium tuberculosis produces pili during human infection. Proc. Natl. Acad. Sci. USA 2007, 104, 5145–5150. [Google Scholar] [CrossRef] [PubMed]
- Chitale, S.; Ehrt, S.; Kawamura, I.; Fujimura, T.; Shimono, N.; Anand, N.; Lu, S.; Cohen-Gould, L.; Riley, L.W. Recombinant mycobacterium tuberculosis protein associated with mammalian cell entry. Cell. Microbiol. 2001, 3, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Arruda, S.; Bomfim, G.; Knights, R.; Huima-Byron, T.; Riley, L.W. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 1993, 10, 1454–1457. [Google Scholar] [CrossRef]
- Krachler, A.M.; Ham, H.; Orth, K. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by Gram-negative pathogens. Proc. Natl. Acad. Sci. USA 2011, 108, 11614–11619. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, T.; Hofbaur, S.; Tegtmeyer, N.; Huber, S.; Sewald, N.; Wessler, S.; Backert, S.; Rieder, G. Helicobacter pylori CagL dependent induction of gastrin expression via a novel αvβ5-integrin–integrin linked kinase signalling complex. Gut 2012, 61, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Ishijima, N.; Suzuki, M.; Ashida, H.; Ichikawa, Y.; Kanegae, Y.; Saito, I.; Borén, T.; Haas, R.; Sasakawa, C.; Mimuro, H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J. Biol. Chem. 2011, 286, 25256–25264. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Claes, I.; Tytgat, H.L.P.; Verhoeven, T.L.A.; Marien, E.; von Ossowski, I.; Reunanen, J.; Palva, A.; de Vos, W.M.; de Keersmaecker, S.C.J.; et al. Functional analysis of lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 2012, 78, 185–193. [Google Scholar] [CrossRef]
- Reunanen, J.; von Ossowski, I.; Hendrickx, A.P.A.; Palva, A.; de Vos, W.M. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2012, 78, 2337–2344. [Google Scholar] [CrossRef] [PubMed]
- Crawford, R.W.; Reeve, K.E.; Gunn, J.S. Flagellated but not hyperfimbriated Salmonella enterica serovar typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 2010, 192, 2981–2990. [Google Scholar] [CrossRef] [PubMed]
- Chessa, D.; Winter, M.G.; Jakomin, M.; Bäumler, A.J. Salmonella enterica serotype typhimurium Std fimbriae bind terminal α(1,2)fucose residues in the cecal mucosa. Mol. Microbiol. 2009, 71, 846–875. [Google Scholar]
- Hase, K.; Kawano, K.; Nochi, T.; Pontes, G.S.; Fukuda, S.; Ebisawa, M.; Kadokura, K.; Tobe, T.; Fujimura, Y.; Kawano, S.; et al. Uptake through glycoprotein 2 of FimH+ bacteria by M cells initiates mucosal immune response. Nature 2009, 462, 226–230. [Google Scholar] [CrossRef]
- Hultgren, S.J.; Normark, S.; Abraham, S.N. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 1991, 45, 383–415. [Google Scholar] [CrossRef] [PubMed]
- Stathopoulos, C.; Hendrixson, D.R.; Thanassi, D.G.; Hultgren, S.J.; St. Geme, J.W., III; Curtiss, R., III. Secretion of virulence determinants by the general secretory pathway in Gram-negative pathogens: An evolving story. Microbes Infect. 2000, 2, 1061–1072. [Google Scholar]
- Phan, G.; Remaut, H.; Wang, T.; Allen, W.J.; Pirker, K.F.; Lebedev, A.; Henderson, N.S.; Geibel, S.; Volkan, E.; Yan, J.; et al. Crystal structure of the FimD usher bound to its cognate FimC–FimH substrate. Nature 2011, 474, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Zakrisson, J.; Wiklund, K.; Axner, O.; Andersson, M. The shaft of the type 1 fimbriae regulates an external force to match the FimH catch bond. Biophys. J. 2013, 104, 2137–2148. [Google Scholar] [CrossRef] [PubMed]
- Carbonnelle, E.; Hélaine, S.; Prouvensier, L.; Nassif, X.; Pelicic, V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 2005, 55, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Siewering, K.; Jain, S.; Friedrich, C.; Webber-Birungi, M.T.; Semchonok, D.A.; Binzen, I.; Wagner, A.; Huntley, S.; Kahnt, J.; Klingl, A.; et al. Peptidoglycan-binding protein TsaP functions in surface assembly of type IV pili. Proc. Natl. Acad. Sci. USA 2014, 111, E953–E961. [Google Scholar] [CrossRef] [PubMed]
- Wolfgang, M.; Park, H.S.; Hayes, S.F.; van Putten, J.P.M.; Koomey, M. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 1998, 95, 14973–14978. [Google Scholar] [CrossRef] [PubMed]
- Maier, B.; Koomey, M.; Sheetz, M.P. A force-dependent switch reverses type IV pilus retraction. Proc. Natl. Acad. Sci. USA 2004, 101, 10961–10966. [Google Scholar] [CrossRef] [PubMed]
- Imhaus, A.F.; Duménil, G. The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J. 2014, 33, 1767–1783. [Google Scholar] [CrossRef] [PubMed]
- Mikaty, G.; Soyer, M.; Mairey, E.; Henry, N.; Dyer, D.; Forest, K.T.; Morand, P.; Guadagnini, S.; Prévost, M.C.; Nassif, X.; et al. Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog. 2009, 5, e1000314. [Google Scholar] [CrossRef] [PubMed]
- Hufnagel, D.A.; Tükel, Ç.; Chapman, M.R. Disease to dirt: The biology of microbial amyloids. PLoS Pathog. 2013, 9, e1003740. [Google Scholar] [CrossRef] [PubMed]
- Goyal, P.; Krasteva, P.V.; van Gerven, N.; Gubellini, F.; van den Broeck, I.; Troupiotis-Tsailaki, A.; Jonckheere, W.; Pehau-Arnaudet, G.; Pinkner, J.S.; Chapman, M.R.; et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 2014, 516, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Cappello, R.; Nataro, J.P. Autotransporter proteins, evolution and redefining protein secretion. Trends Microbiol. 2000, 8, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Totsika, M.; Wells, T.J.; Beloin, C.; Valle, J.; Allsopp, L.P.; King, N.P.; Ghigo, J.M.; Schembri, M.A. Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 2179–2189. [Google Scholar] [CrossRef] [PubMed]
- Bölin, I.; Wolf-Watz, H. Molecular cloning of the temperature-inducible outer membrane protein 1 of Yersinia pseudotuberculosis. Infect. Immun. 1984, 43, 72–78. [Google Scholar] [PubMed]
- El-Kirat-Chatel, S.; Mil-Homens, D.; Beaussart, A.; Fialho, A.M.; Dufrêne, Y.F. Single-molecule atomic force microscopy unravels the binding mechanism of a Burkholderia cenocepacia trimeric autotransporter adhesin. Mol. Microbiol. 2013, 89, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xie, J. Mammalian cell entry gene family of Mycobacterium tuberculosis. Mol. Cell Biochem. 2011, 352, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, M.D.L.P.; Klepp, L.; Nuñez-García, J.; Blanco, F.C.; Soria, M.; García-Pelayo, M.d.C.; Bianco, M.V.; Cataldi, A.A.; Golby, P.; Jackson, M.; et al. Mce3R, a TetR-type transcriptional repressor, controls the expression of a regulon involved in lipid metabolism in Mycobacterium tuberculosis. Microbiology 2009, 155, 2245–2255. [Google Scholar] [CrossRef] [PubMed]
- Casali, N.; Riley, L. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 2007, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chandolia, A.; Chaudhry, U.; Brahmachari, V.; Bose, M. Comparison of mammalian cell entry operons of mycobacteria: In silico analysis and expression profiling. FEMS Immunol. Med. Microbiol. 2005, 43, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Krachler, A.M.; Orth, K. Functional characterization of the interaction between bacterial adhesin multivalent adhesion molecule 7 (MAM7) protein and its host cell ligands. J. Biol. Chem. 2011, 286, 38939–38947. [Google Scholar] [CrossRef] [PubMed]
- Thinwa, J.; Segovia, J.A.; Bose, S.; Dube, P.H. Integrin-mediated first signal for inflammasome activation in intestinal epithelial cells. J. Immunol. 2014, 193, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Toller, I.M.; Neelsen, K.J.; Steger, M.; Hartung, M.L.; Hottiger, M.O.; Stucki, M.; Kalali, B.; Gerhard, M.; Sartori, A.A.; Lopes, M.; et al. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA 2011, 108, 14944–14949. [Google Scholar] [CrossRef]
- Young, B.P.; Shin, J.J.H.; Orij, R.; Chao, J.T.; Li, S.C.; Guan, X.L.; Khong, A.; Jan, E.; Wenk, M.R.; Prinz, W.A.; et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 2010, 329, 1085–1088. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Devaiah, S.P.; Zhang, W.; Welti, R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006, 45, 250–278. [Google Scholar] [CrossRef] [PubMed]
- Kooijman, E.E.; Chupin, V.; de Kruijff, B.; Burger, K.N.J. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 2003, 4, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Andresen, B.T.; Rizzo, M.A.; Shome, K.; Romero, G. The role of phosphatidic acid in the regulation of the Ras/MEK/Erk signaling cascade. FEBS Lett. 2002, 531, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Vilella-Bach, M.; Bachmann, R.; Flanigan, A.; Chen, J. Phosphatidic acid-mediated mitogenic activation of mtor signaling. Science 2001, 294, 1942–1945. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Stones, D.H.; Hawley, C.A.; Watson, C.A.; Krachler, A.M. Multivalent adhesion molecule 7 clusters act as signaling platform for host cellular GTPase activation and facilitate epithelial barrier dysfunction. PLoS Pathog. 2014, 10, e1004421. [Google Scholar] [CrossRef] [PubMed]
- Tükel, Ç.; Wilson, R.P.; Nishimori, J.H.; Pezeshki, M.; Chromy, B.A.; Bäumler, A.J. Responses to amyloids of microbial and host origin are mediated through Toll-like receptor 2. Cell Host Microbe 2009, 6, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Ofek, I.; Hasty, D.L.; Sharon, N. Anti-adhesion therapy of bacterial diseases: Prospects and problems. FEMS Immunol. Med. Microbiol. 2003, 38, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Krachler, A.M.; Orth, K. Made to stick: Anti-adhesion therapy for bacterial infections. Microbe Magazine 2013. [Google Scholar]
- Hartmann, M.; Papavlassopoulos, H.; Chandrasekaran, V.; Grabosch, C.; Beiroth, F.; Lindhorst, T.K.; Röhl, C. Inhibition of bacterial adhesion to live human cells: Activity and cytotoxicity of synthetic mannosides. FEBS Lett. 2012, 586, 1459–1465. [Google Scholar] [CrossRef] [PubMed]
- Totsika, M.; Kostakioti, M.; Hannan, T.J.; Upton, M.; Beatson, S.A.; Janetka, J.W.; Hultgren, S.J.; Schembri, M.A. A FimH inhibitor prevents acute bladder infection and treats chronic cystitis caused by multidrug resistant uropathogenic Escherichia coli ST131. J. Infect. Dis. 2013, 208, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Krachler, A.M.; Ham, H.; Orth, K. Turnabout is fair play. Virulence 2012, 3, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Krachler, A.M.; Mende, K.; Murray, C.; Orth, K. In vitro characterization of multivalent adhesion molecule 7-based inhibition of multidrug-resistant bacteria isolated from wounded military personnel. Virulence 2012, 3, 389–399. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stones, D.H.; Krachler, A.-M. Fatal Attraction: How Bacterial Adhesins Affect Host Signaling and What We Can Learn from Them. Int. J. Mol. Sci. 2015, 16, 2626-2640. https://doi.org/10.3390/ijms16022626
Stones DH, Krachler A-M. Fatal Attraction: How Bacterial Adhesins Affect Host Signaling and What We Can Learn from Them. International Journal of Molecular Sciences. 2015; 16(2):2626-2640. https://doi.org/10.3390/ijms16022626
Chicago/Turabian StyleStones, Daniel H., and Anne-Marie Krachler. 2015. "Fatal Attraction: How Bacterial Adhesins Affect Host Signaling and What We Can Learn from Them" International Journal of Molecular Sciences 16, no. 2: 2626-2640. https://doi.org/10.3390/ijms16022626





