Quantification and Application of Potential Epigenetic Markers in Maternal Plasma of Pregnancies with Hypertensive Disorders
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Clinical Characteristics
| Characteristics | Controls (n = 188) | EO-PE (n = 16) | LO-PE (n = 47) | GH (n = 29) | P a | P b | P c | P d | P e | P f |
|---|---|---|---|---|---|---|---|---|---|---|
| Maternal age (years) | 33.0 (31.0–36.0) | 34.0 (31.6–36.0) | 34.1 (33.0–36.1) | 35.0 (33.0–37.0) | 0.204 | 0.061 | 0.059 | 0.383 | 0.393 | 0.809 |
| BMI (kg/m2) | 20.2 (18.8–21.9) | 22.3 (22.1–29.4) | 22.6 (20.3–23.5) | 22.8 (21.2–30.5) | 0.000 | 0.000 | 0.000 | 0.073 | 0.948 | 0.044 |
| Nulliparity | 76 (40.4) | 6 (37.5) | 24 (51.1) | 16 (55.2) | 0.819 | 0.187 | 0.135 | 0.397 | 0.353 | 0.815 |
| Alcohol intake | 44 (23.4) | 6 (37.5) | 13 (27.7) | 5 (17.2) | 0.229 | 0.543 | 0.460 | 0.534 | 0.161 | 0.408 |
| Tobacco use | 2 (1.1) | – | – | – | – | – | – | – | – | – |
| Maximum SBP (mmHg) | 109.0 (101.0–118.0) | 162.0 (160.0–173.0) | 147.0 (140.5–150.5) | 157.0 (150.5–167.0) | 0.000 | 0.000 | 0.000 | 0.000 | 0.387 | 0.000 |
| Maximum DBP (mmHg) | 63.0 (57.0–69.0) | 99.0 (93.0–100.0) | 90.0 (86.0–94.0) | 96.0 (89.0–98.5) | 0.000 | 0.000 | 0.000 | 0.035 | 0.355 | 0.077 |
| Birthweight (g) | 3290.0 (3040.0–3495.0) | 2265.0 (1878.8–2362.5) | 2920.0 (2572.5–3297.5) | 3012.5 (2800.0–3290.0) | 0.000 | 0.000 | 0.001 | 0.002 | 0.000 | 0.384 |
| Sex ratio of fetus (male:female) | 92:96 | 10:6 | 16:31 | 11:18 | 0.298 | 0.067 | 0.269 | 0.076 | 0.133 | 0.807 |
| GA at delivery (weeks) | 39.5 (39.1–40.3) | 35.8 (34.4–36.4) | 38.3 (36.8–39.3) | 38.6 (38.2–39.5) | 0.000 | 0.000 | 0.006 | 0.003 | 0.000 | 0.032 |
| GA at sampling (weeks) | – | – | – | – | – | – | – | – | – | – |
| First trimester | 12.1 (8.4–12.4) | 12.0 (11.1–12.3) | 12.3 (11.7–12.5) | 12.3 (12.2–12.3) | 0.850 | 0.382 | 0.205 | 0.571 | 0.567 | 0.970 |
| Sceond trimester | 21.2 (16.4–25.0) | 20.9 (16.6–25.2) | 18.0 (16.6–24.0) | 17.9 (16.3–24.4) | 0.722 | 0.060 | 0.090 | 0.451 | 0.189 | 0.903 |
| Third trimester | 36.5 (36.1–37.0) | 36.3 (35.3–36.6) | 36.9 (36.2–38.2) | 36.6 (36.6–37.7) | 0.063 | 0.329 | 0.100 | 0.187 | 0.073 | 0.967 |
2.1.2. Selection of Fetal-Specific Hypermethylated CpG loci by High-resolution Tiling Array Analysis
| Target Regions | Chr. | Description | Probe Position | Placenta (Log2) | Normal Blood (Log2) | ||
|---|---|---|---|---|---|---|---|
| Start | Stop | Pregnant | Non-Pregnant | ||||
| SOD1 | 21 | Inside | 33037889 | 33037933 | 2.62 ± 0.53 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| DSCR3 | 21 | Inside | 38629500 | 38629559 | 2.77 ± 0.43 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| C2CD2 | 21 | Inside | 43316160 | 43316204 | 3.45 ± 1.02 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| UMODL1 | 21 | Inside | 43483898 | 43483942 | 3.14 ± 0.84 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| ENST00000433952: -7924 | 21 | Promoter | 40357602 | 40357646 | 3.39 ± 0.77 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| ENST00000433952: -7988 | 21 | Promoter | 40357666 | 40357710 | 3.52 ± 1.12 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| ENST00000450830: -4439 | 21 | Promoter | 34524477 | 34524521 | 2.85 ± 0.82 | 0.00 ± 0.00 | 0.00 ± 0.00 |

2.1.3. Confirmation of High-Resolution Tiling Array Data Using Bisulfite Direct Sequencing
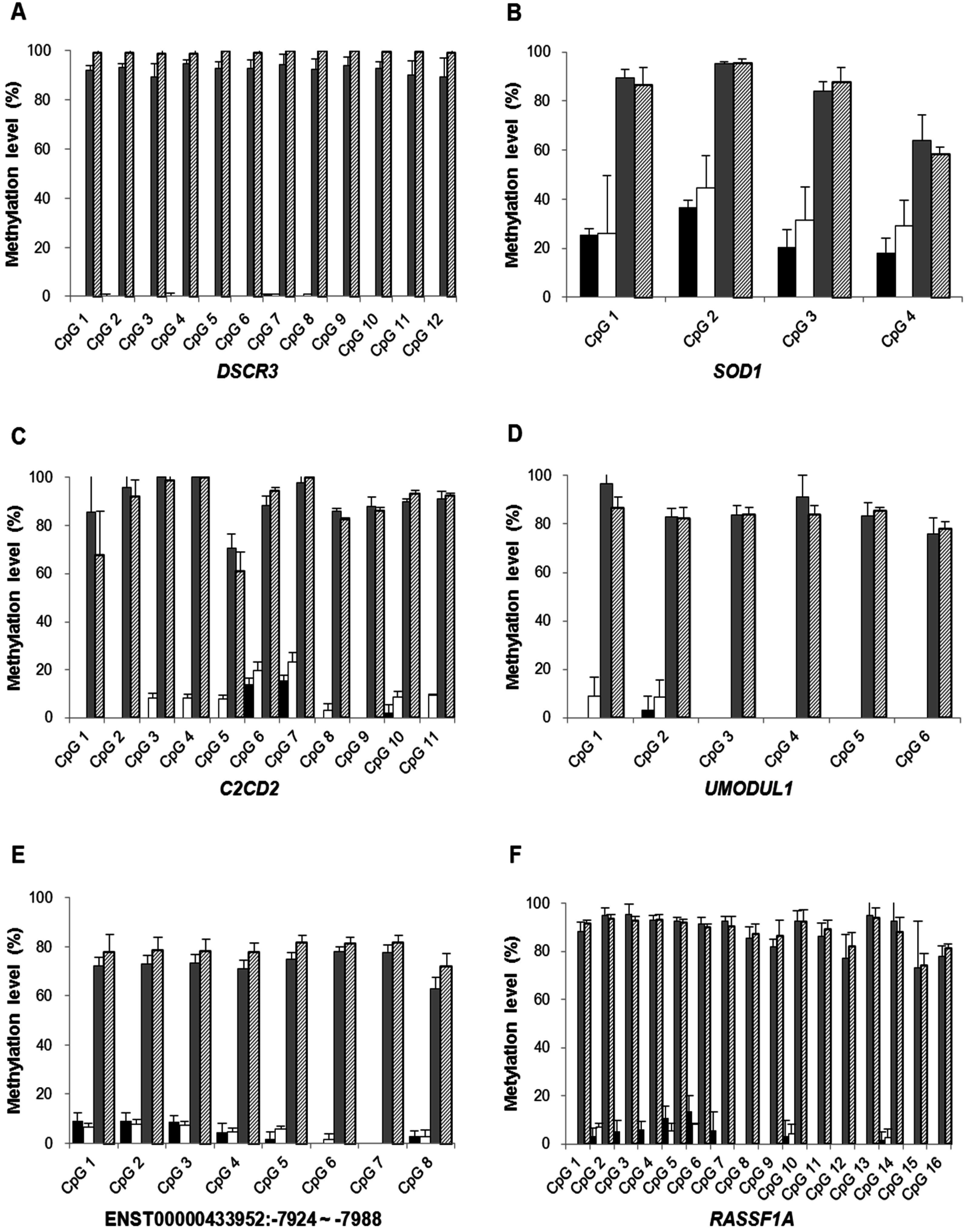
2.1.4. Quantification of DSCR3, RASSF1A, SRY and GADH in Maternal Plasma of Normal Pregnancies
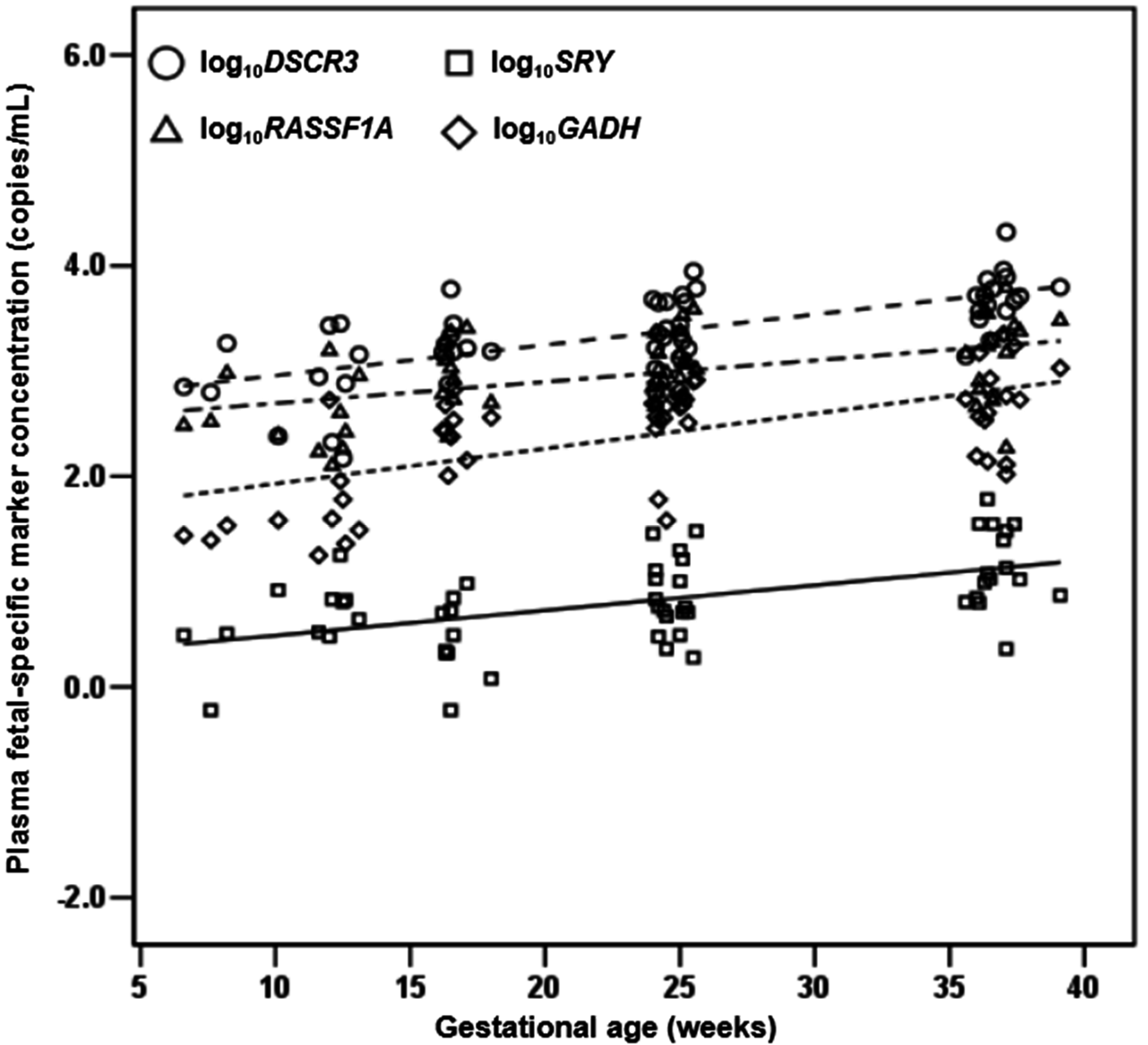
2.1.5. DSCR3 and RASSF1A Concentrations in Maternal Plasma between Normal Pregnancies and Pregnancies with Hypertensive Disorders
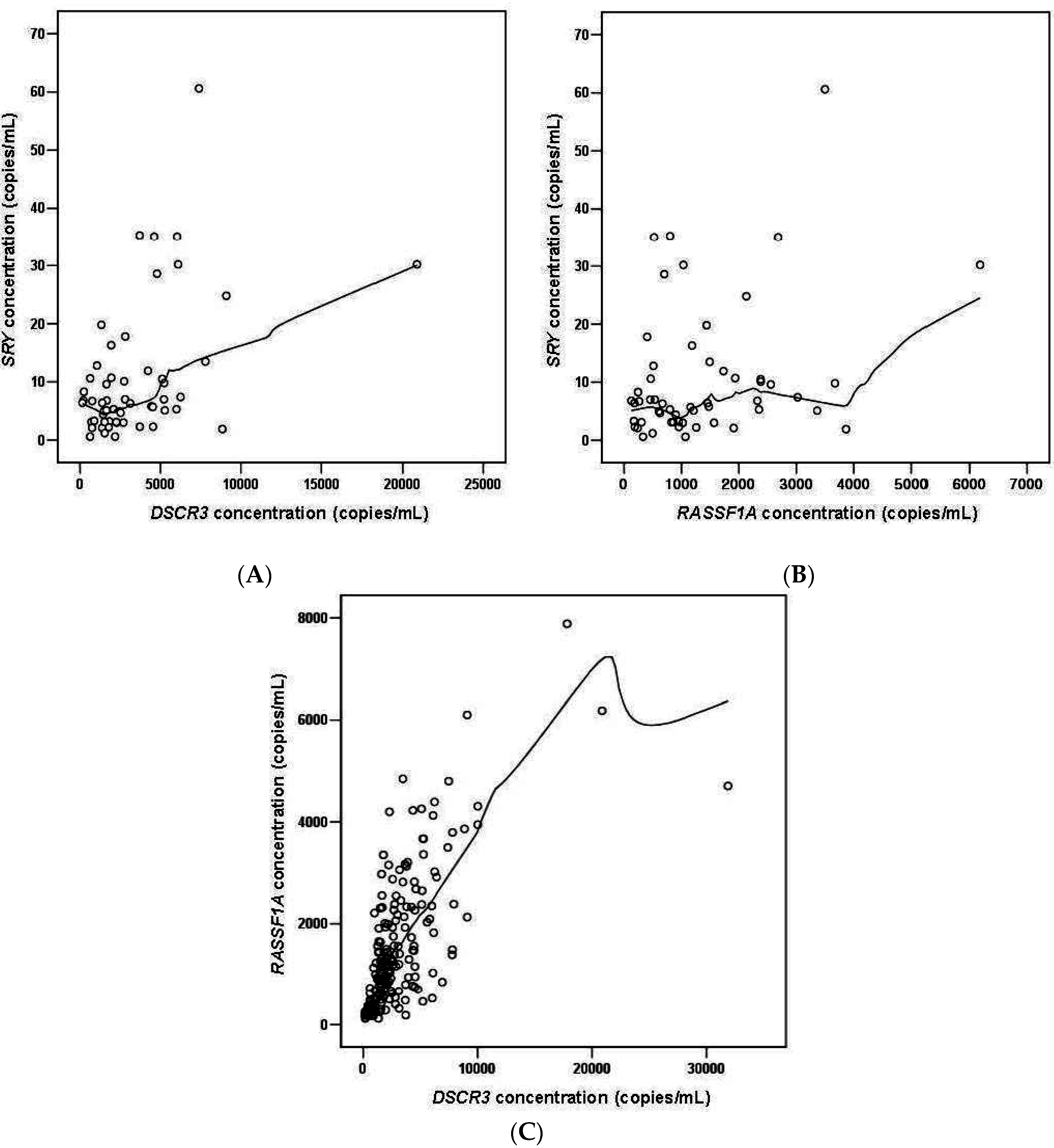
2.2. Discussion
| Marker | Controls | EO-PE | LO-PE | GH | P a | P b | P c | P d | P e | P f |
|---|---|---|---|---|---|---|---|---|---|---|
| 6–14 weeks | (n = 52) | (n = 4) | (n = 10) | (n = 10) | – | – | – | – | – | – |
| DSCR3 | 890.9 (538.9–1530.0) | 1007.7 (721.5–1454.3) | 939.1 (699.6–1718.2) | 1098.3 (561.7–2073.6) | 0.818 | 0.467 | 0.566 | 0.945 | 0.733 | 0.739 |
| RASSF1A | 445.6 (262.9–936.4) | 858.0 (652.6–1215.2) | 595.2 (382.1–807.2) | 450.1 (385.1–719.7) | 0.118 | 0.503 | 0.444 | 0.374 | 0.188 | 0.853 |
| 15–23 weeks | (n = 31) | (n = 3) | (n = 16) | (n = 7) | – | – | – | – | – | – |
| DSCR3 | 1779.7 (1443.8–2590.6) | 2594.0 (2171.5–3075.6) | 2573.7 (2159.9–2936.4) | 1692.8 (1220.7–2009.1) | 0.289 | 0.121 | 0.580 | 0.958 | 0.183 | 0.076 |
| RASSF1A | 1333.8 (727.9–2403.3) | 2305.5 (1571.0–2842.2) | 1601.4 (1154.4–2036.8) | 1013.3 (716.5–1375.0) | 0.449 | 0.946 | 0.282 | 0.421 | 0.183 | 0.103 |
| 24–32 weeks | (n = 47) | (n = 5) | (n = 7) | (n = 5) | – | – | – | – | – | – |
| DSCR3 | 2521.3 (1824.9–4438.2) | 6772.5 (5251.1–7876.2) | 3295.0 (1118.3–6765.2) | 2725.5 (1372.9–6415.1) | 0.018 | 0.920 | 0.857 | 0.268 | 0.222 | 1.000 |
| RASSF1A | 1228.3 (827.6–2357.1) | 3100.8 (2359.7–4600.0) | 819.1 (610.1–1177.3) | 1765.5 (406.2–2965.0) | 0.032 | 0.212 | 1.000 | 0.149 | 0.222 | 0.639 |
| 33–41 weeks | (n = 58) | (n = 4) | (n = 14) | (n = 7) | – | – | – | – | – | – |
| DSCR3 | 4212.9 (2841.1–6162.5) | 10,632.4 (8426.2–16,027.7) | 11,164.3 (7733.5–13,909.9) | 6985.4 (5070.0–8881.8) | 0.001 | 0.000 | 0.079 | 0.721 | 0.109 | 0.056 |
| RASSF1A | 1777.2 (930.2–2674.1) | 4002.8 (3449.6–5142.4) | 5051.3 (3441.6–6787.2) | 2979.7 (1915.7–3763.8) | 0.008 | 0.000 | 0.173 | 0.721 | 0.109 | 0.016 |
3. Materials and Methods
3.1. Study Participants and Samples
3.2. DNA Preparation
3.3. Data Analysis of High-Resolution Tiling Arrays
3.4. Bisulfite Direct Sequencing
3.5. Methylation Quantification of Bisulfite Direct Sequencing Data
3.6. Enrichment of Methylated DNA
3.7. Quantitative Real-Time PCR
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sibai, B.M.; Stella, C.L. Diagnosis and management of atypical preeclampsia-eclampsia. Am. J. Obstet. Gynecol. 2009. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Lindheimer, M.D.; de Swiet, M.; van Assche, A.; Moutquin, J.M. The classification and diagnosis of the hypertensive disorders of pregnancy: Statement from the international society for the Study of hypertension in pregnancy (ISSHP). Hypertens. Pregnancy 2001, 20, 9–14. [Google Scholar] [CrossRef]
- Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lim, J.H.; Park, S.Y.; Kim, M.Y.; Choi, J.S.; Ryu, H.M. Non-invasive prenatal determination of fetal gender using QF-PCR analysis of cell-free fetal DNA in maternal plasma. Clin. Chim. Acta 2012, 413, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Geifman-Holtzman, O.; Grotegut, C.A.; Gaughan, J.P. Diagnostic accuracy of noninvasive fetal Rh genotyping from maternal blood e a meta-analysis. Am. J. Obstet. Gynecol. 2006, 195, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.; Lun, F.M.; Chan, K.C.; Tsui, N.B.; Chong, K.C.; Lau, T.K.; Leung, T.Y.; Zee, B.C.; Cantor, C.R.; Chiu, R.W. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc. Natl. Acad. Sci. USA 2007, 104, 13116–13121. [Google Scholar] [CrossRef] [PubMed]
- Leung, T.N.; Zhang, J.; Lau, T.K.; Chan, L.Y.; Lo, Y.M. Increased maternal plasma fetal DNA concentrations in women who eventually develop preeclampsia. Clin. Chem. 2001, 47, 137–139. [Google Scholar] [PubMed]
- Caramelli, E.; Rizzo, N.; Concu, M.; Simonazzi, G.; Carinci, P.; Bondavalli, C.; Bovicelli, L.; Farina, A. Cell-free fetal DNA concentration in plasma of patients with abnormal uterine artery Doppler waveform and intrauterine growth restriction—A pilot study. Prenat. Diagn. 2003, 23, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.; Ding, C.; Gerovassili, A.; Yeung, S.W.; Chiu, R.W.; Leung, T.N.; Lau, T.K.; Chim, S.S.; Chung, G.T.; Nicolaides, K.H.; et al. Hypermethylated RASSF1A in maternal plasma: A universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin. Chem. 2006, 52, 2211–2218. [Google Scholar] [CrossRef] [PubMed]
- Papantoniou, N.; Bagiokos, V.; Agiannitopoulos, K.; Kolialexi, A.; Destouni, A.; Tounta, G.; Kanavakis, E.; Antsaklis, A.; Mavrou, A. RASSF1A in maternal plasma as a molecular marker of preeclampsia. Prenat. Diagn. 2013, 33, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wang, J.; Liu, R.; Yang, J.; Cui, K.; Wu, Y.; Guo, J.; Mu, Y.; Wang, X. Quantification and application of the placental epigenetic signature of the RASSF1A gene in maternal plasma. Prenat. Diagn. 2010, 30, 778–782. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kim, S.Y.; Park, S.Y.; Ahn, H.K.; Chung, J.H.; Ryu, H.M. Association of fetal-derived hypermethylated RASSF1A concentration in placenta-mediated pregnancy complications. Placenta 2013, 34, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Yuen, R.K.; Peñaherrera, M.S.; von Dadelszen, P.; McFadden, D.E.; Robinson, W.P. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur. J. Hum. Genet. 2010, 18, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.D.; Yuen, R.K.; Lim, B.K.; McFadden, D.E.; von Dadelszen, P.; Robinson, W.P. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Mol. Hum. Reprod. 2013, 19, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Sitras, V.; Paulssen, R.H.; Grønaas, H.; Leirvik, J.; Hanssen, T.A.; Vårtun, A.; Acharya, G. Differential placental gene expression in severe preeclampsia. Placenta 2009, 30, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, H.; Ota, S.; Suzuki, M.; Kato, T.; Sekiya, T.; Kurahashi, H.; Udagawa, Y. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod. Biol. Endocrinol. 2011, 9, 107. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Lee, D.E.; Park, S.Y.; Kim, D.J.; Ahn, H.K.; Han, Y.J.; Kim, M.Y.; Ryu, H.M. Disease specific characteristics of fetal epigenetic markers for non-invasive prenatal testing of trisomy 21. BMC Med. Genom. 2014, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Cotter, A.M.; Martinm, C.M.; O'leary, J.J.; Daly, S.F. Increased fetal DNA in the maternal circulation in early pregnancy is associated with an increased risk of preeclampsia. Am. J. Obstet. Gynecol. 2004, 191, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Crowley, A.; Martin, C.; Fitzpatrick, P.; Sheils, O.; O'Herlihy, C.; O'Leary, J.J.; Byrne, B.M. Free fetal DNA is not increased before 20 weeks in intrauterine growth restriction or pre-eclampsia. Prenat. Diagn. 2007, 27, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.C.; Musci, T.; Song, K.; Syngelaki, A.; Nicolaides, K.H. Maternal plasma cell-free fetal and maternal DNA at 11–13 weeks’ gestation: Relation to fetal and maternal characteristics and pregnancy outcomes. Fetal Diagn. Ther. 2013, 33, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.J.; Qian, C.; Leshane, E.S.; Yu, K.F.; England, L.J.; Schisterman, E.F.; Wataganara, T.; Romero, R.; Bianchi, D.W. Two-stage elevation of cell-free fetal DNA in maternal sera before onset of preeclampsia. Am. J. Obstet. Gynecol. 2004, 190, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Sifakis, S.; Zaravinos, A.; Maiz, N.; Spandidos, D.A.; Nicolaides, K.H. First-trimester maternal plasma cell-free fetal DNA and preeclampsia. Am. J. Obstet. Gynecol. 2009. [Google Scholar] [CrossRef] [PubMed]
- Illanes, S.; Parra, M.; Serra, R.; Pino, K.; Figueroa-Diesel, H.; Romero, C.; Arraztoa, J.A.; Michea, L.; Soothill, P.W. Increased free fetal DNA levels in early pregnancy plasma of women who subsequently develop preeclampsia and intrauterine growth restriction. Prenat. Diagn. 2009, 29, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Goswami, D.; Tannetta, D.S.; Magee, L.A.; Fuchisawa, A.; Redman, C.W.; Sargent, I.L.; von Dadelszen, P. Excess syncytiotrophoblast microparticle shedding is a feature of early-onset pre-eclampsia, but not normotensive intrauterine growth restriction. Placenta 2006, 27, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Oudejans, C.B.; van Dijk, M.; Oosterkamp, M.; Lachmeijer, A.; Blankenstein, M.A. Genetics of preeclampsia: Paradigm shifts. Hum. Genet. 2007, 120, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Redman, C.W.; Sargent, I.L. Latest advances in understanding preeclampsia. Science 2005, 308, 1592–1594. [Google Scholar] [CrossRef] [PubMed]
- Correa, R.R.; Gilio, D.B.; Cavellani, C.L.; Paschoini, M.C.; Oliveira, F.A.; Peres, L.C.; Reis, M.A.; Teixeira, V.P.; Castro, E.C. Placental morphometrical and histopathology changes in the different clinical presentations of hypertensive syndromes in pregnancy. Arch. Gynecol. Obstet. 2008, 277, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Maloney, K.F.; Heller, D.; Baergen, R.N. Types of maternal hypertensive disease and their association with pathologic lesions and clinical factors. Fetal Pediatr. Pathol. 2012, 31, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Noori, M.; Donald, A.E.; Angelakopoulou, A.; Hingorani, A.D.; Williams, D.J. Prospective study of placental angiogenic factors and maternal vascular function before and after preeclampsia and gestational hypertension. Circulation 2010, 122, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhang, J.; Li, Q.; Zhou, X.; Wang, T.; Xu, M.; Xia, S.; Xing, Q.; Wang, L.; He, L.; et al. DNA methylome profiling of maternal peripheral blood and placentas reveal potential fetal DNA markers for non-invasive prenatal testing. Mol. Hum. Reprod. 2014, 20, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Aquilina, J.; Barnett, A.; Thompson, O.; Harrington, K. Second-trimester maternal serum inhibin A concentrations as an early marker for preeclampsia. Am. J. Obstet. Gynecol. 1999, 181, 131–136. [Google Scholar] [CrossRef]
- Dugoff, L.; Hobbins, J.C.; Malone, F.D.; Vidaver, J.; Sullivan, L.; Canick, J.A.; Lambert-Messerlian, G.M.; Porter, T.F.; Luthy, D.A.; Comstock, C.H.; et al. FASTER Trial Research Consortium. Quad screen as a predictor of adverse pregnancy outcome. Obstet. Gynecol. 2005, 106, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, B.; Levytska, K.; Kingdom, J.; Walker, M.; Baczyk, D.; Keating, S. Villous trophoblast abnormalities in extremely preterm deliveries with elevated second trimester maternal serum hCG or inhibin-A. Placenta 2011, 32, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Goetzinger, K.R.; Odibo, A.O. Screening for abnormal placentation and adverse pregnancy outcomes with maternal serum biomarkers in the second trimester. Prenat. Diagn. 2014, 34, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, Y.; Fei, J.; Chang, X.; Fan, W.; Qian, X.; Zhang, T.; Lu, D. Rapid quantification of DNA methylation by measuring relative peak heights in direct bisulfite-PCR sequencing traces. Lab. Investig. 2010, 90, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.; Tein, M.S.; Lau, T.K.; Haines, C.J.; Leung, T.N.; Poon, P.M.; Wainscoat, J.S.; Johnson, P.J.; Chang, A.M.; Hjelm, N.M. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am. J. Hum. Genet. 1998, 62, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.J.; Kim, S.Y.; Lim, J.H.; Kwak, D.W.; Park, S.Y.; Ryu, H.M. Quantification and Application of Potential Epigenetic Markers in Maternal Plasma of Pregnancies with Hypertensive Disorders. Int. J. Mol. Sci. 2015, 16, 29875-29888. https://doi.org/10.3390/ijms161226201
Kim HJ, Kim SY, Lim JH, Kwak DW, Park SY, Ryu HM. Quantification and Application of Potential Epigenetic Markers in Maternal Plasma of Pregnancies with Hypertensive Disorders. International Journal of Molecular Sciences. 2015; 16(12):29875-29888. https://doi.org/10.3390/ijms161226201
Chicago/Turabian StyleKim, Hyun Jin, Shin Young Kim, Ji Hyae Lim, Dong Wook Kwak, So Yeon Park, and Hyun Mee Ryu. 2015. "Quantification and Application of Potential Epigenetic Markers in Maternal Plasma of Pregnancies with Hypertensive Disorders" International Journal of Molecular Sciences 16, no. 12: 29875-29888. https://doi.org/10.3390/ijms161226201





