New Whitening Constituents from Taiwan-Native Pyracantha koidzumii: Structures and Tyrosinase Inhibitory Analysis in Human Epidermal Melanocytes
Abstract
:1. Introduction
2. Results and Discussion

2.1. New Compounds 9 and 13 Isolated from Pyracantha koidzumii
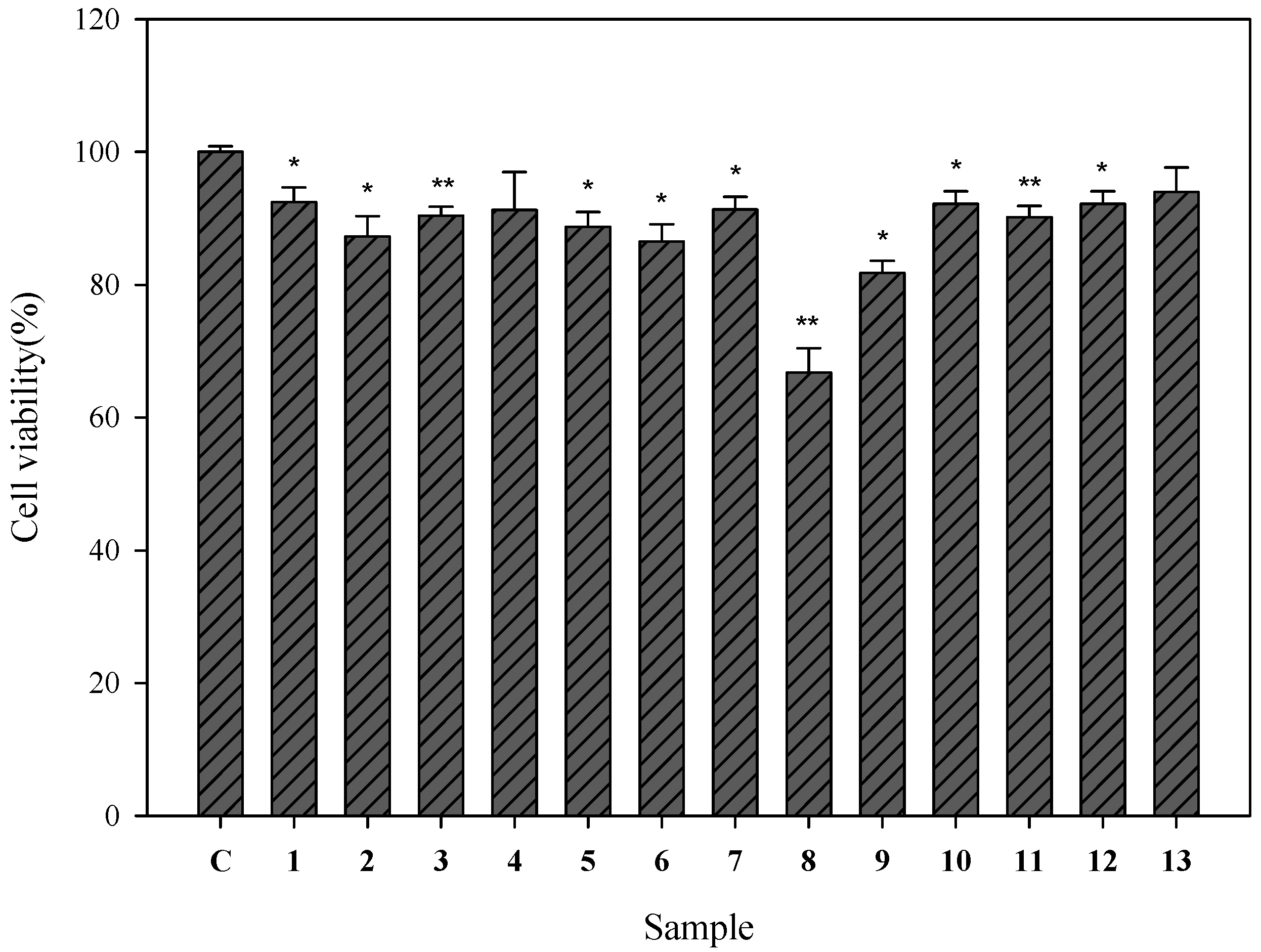
2.2. Cell Viability of Human Epidermal Melanocytes Treated with Compounds Isolated from Pyracantha koidzumii
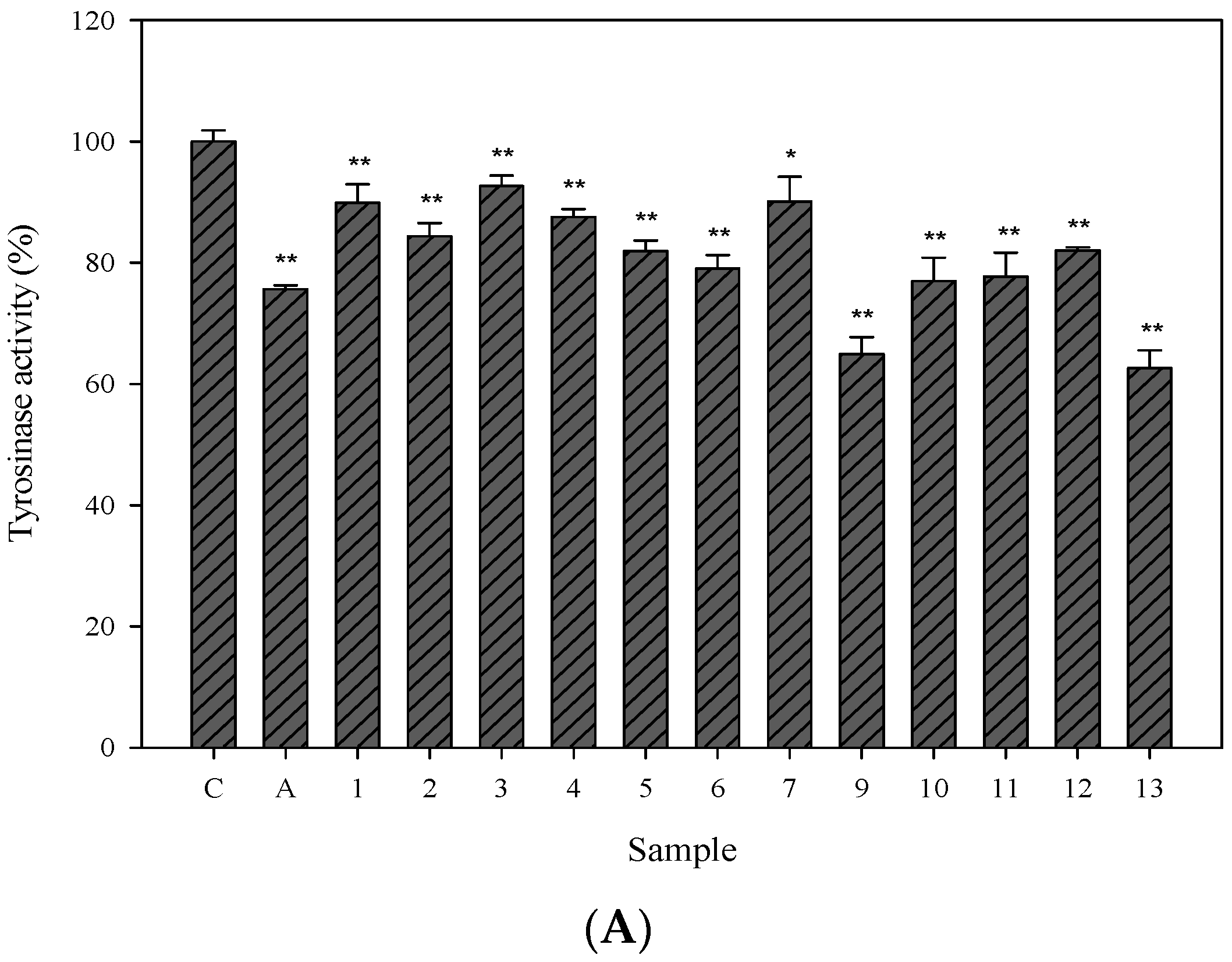
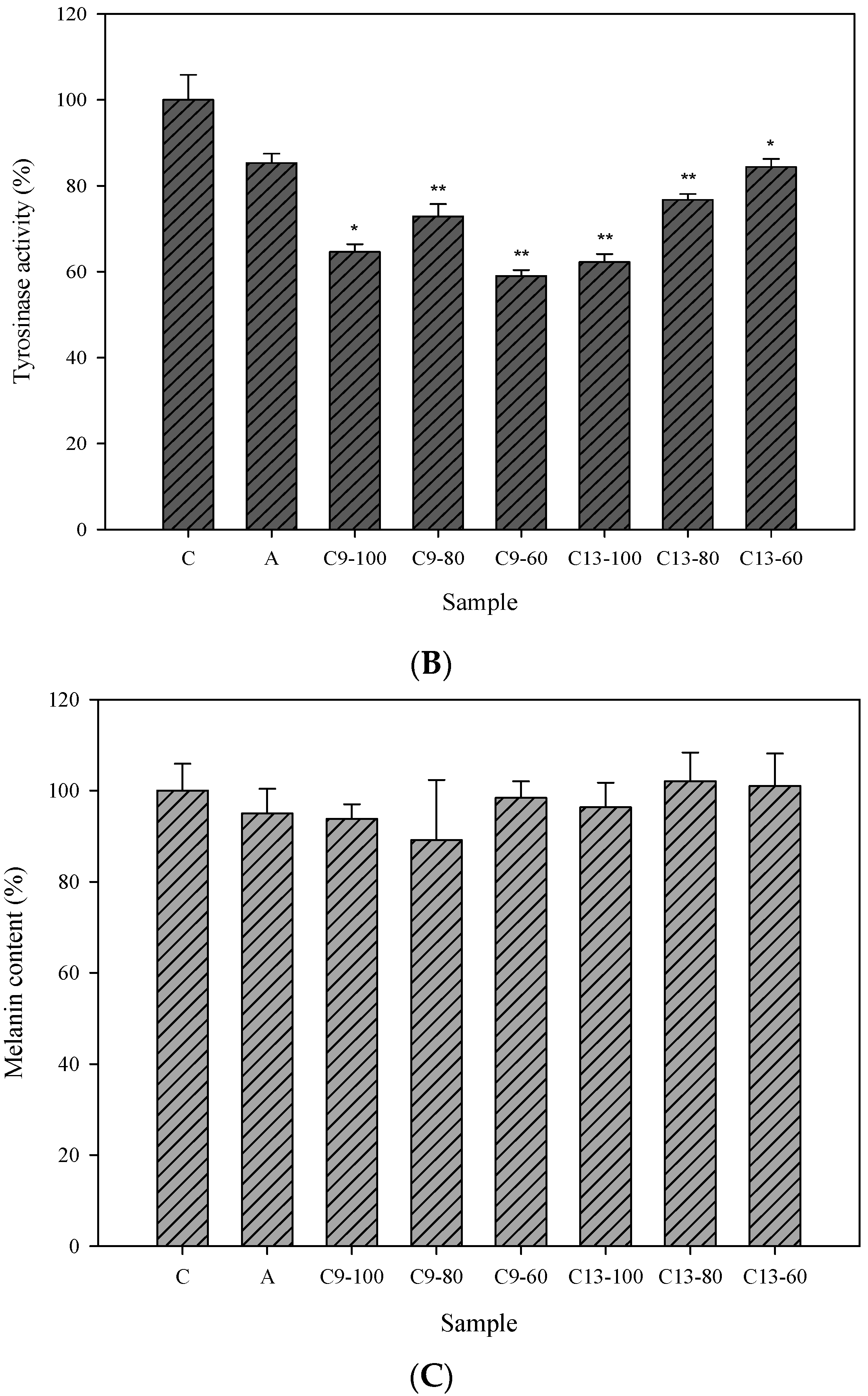
2.3. Cellular Tyrosinase-Inhibitory Activity and Melanin Content of the Isolated Compounds in Human Epidermal Melanocytes
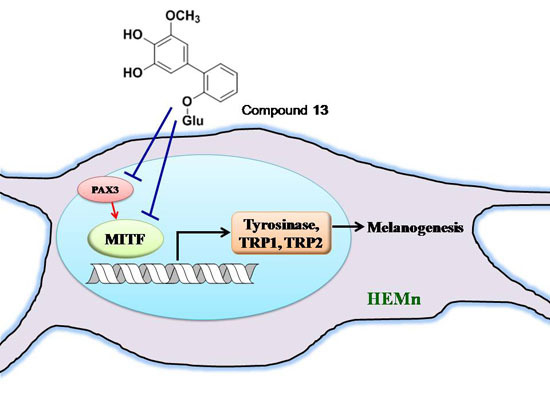
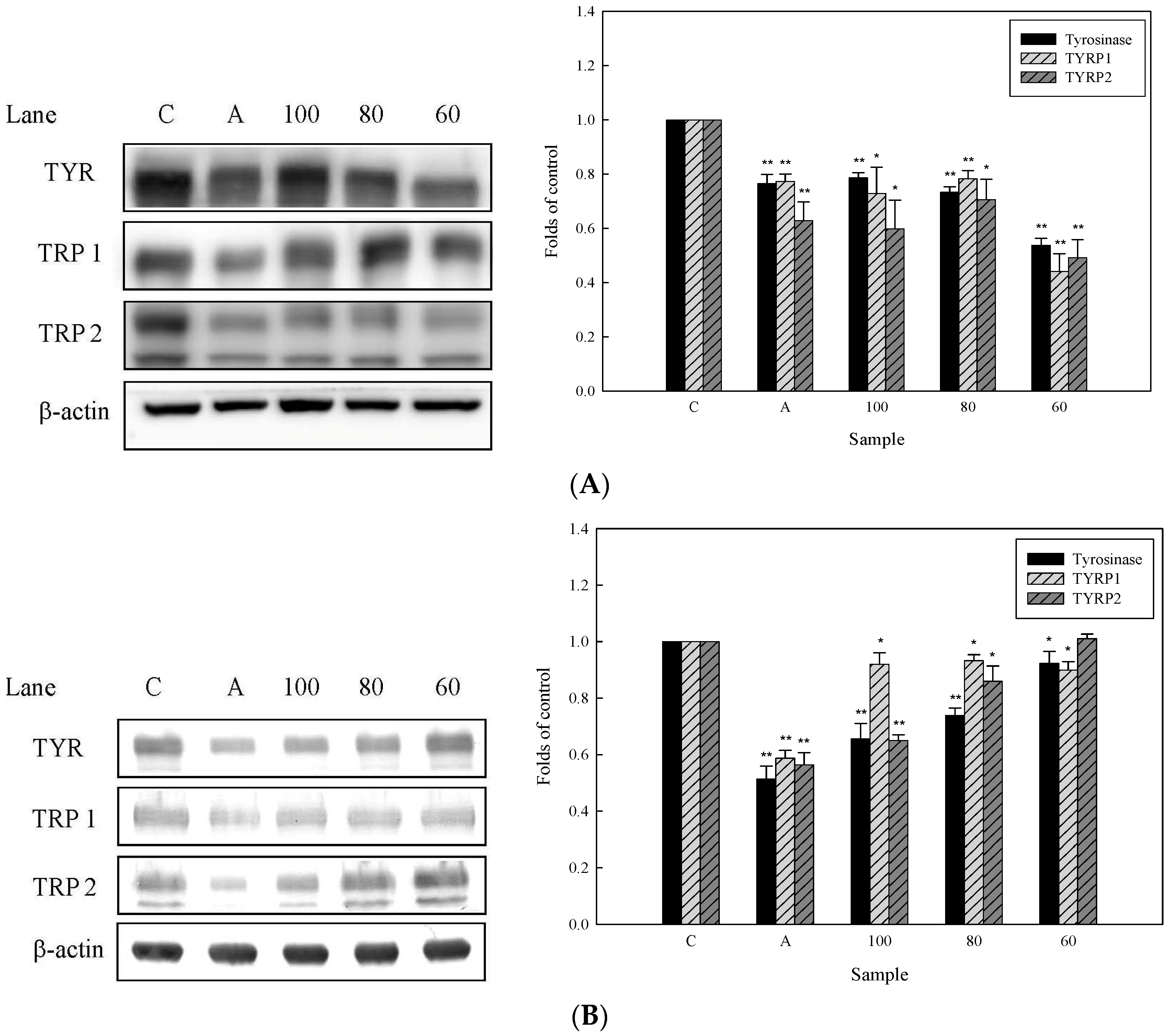
2.4. Effects of 3,6-Dihydroxy-2,4-dimethoxy-dibenzofuran (9) and 3,4-Dihydroxy-5-methoxybiphenyl-2′-O-β-d-glucopyranoside (13) on the Expression of Tyrosinase-Related Proteins in Human Epidermal Melanocytes
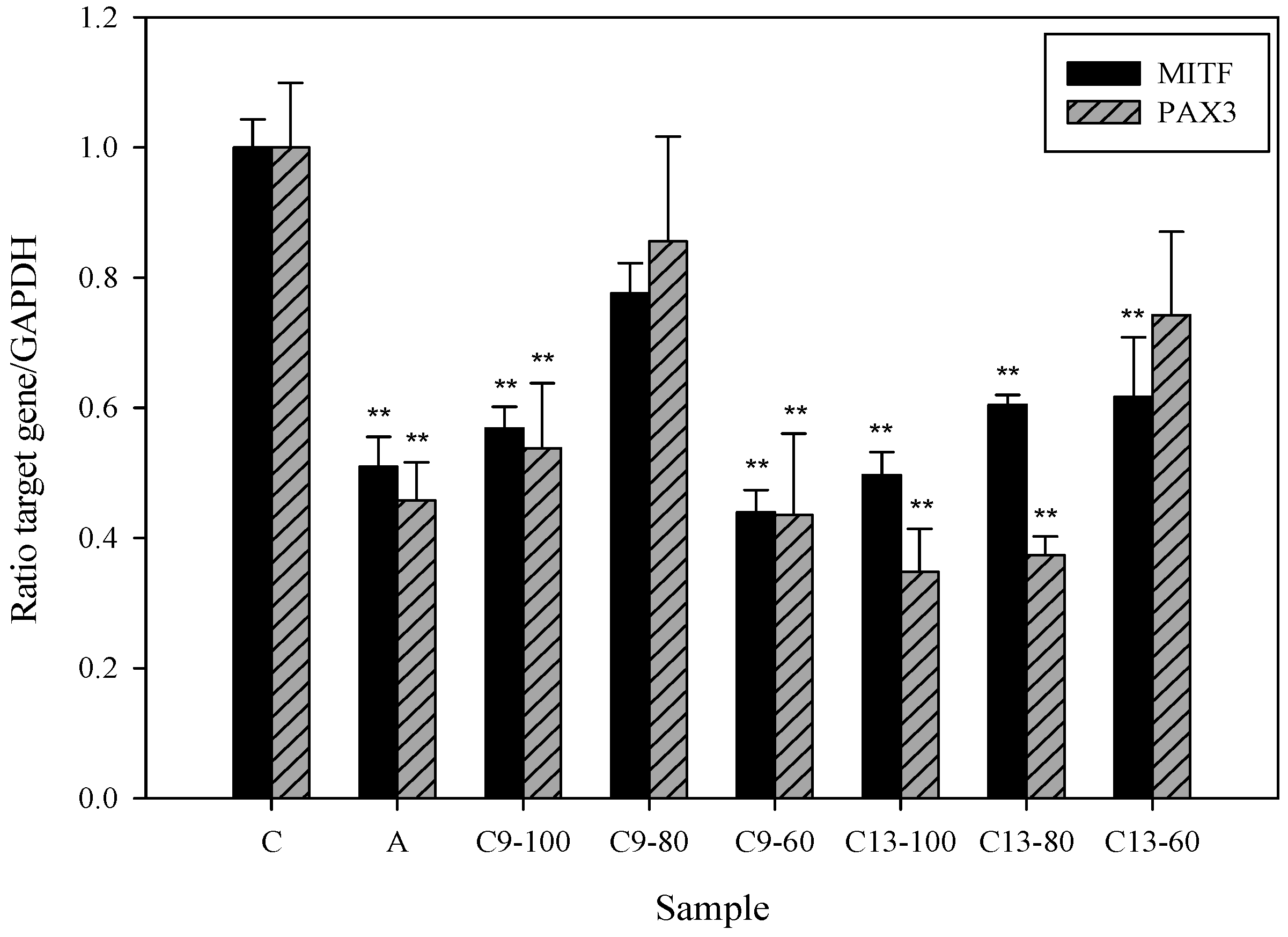
2.5. Effects of 3,6-Dihydroxy-2,4-dimethoxy-dibenzofuran (9) and 3,4-Dihydroxy-5-methoxybiphenyl-2′-O-β-d-glucopyranoside (13) on the Expression of MITF and PAX3 mRNA in Human Epidermal Melanocytes
2.6. Tyrosinase Kinetic Analysis on 3,4-Dihydroxy-5-methoxybiphenyl-2ʹ-O-β-d-glucopyranoside (13) Treatment of Human Epidermal Melanocytes

3. Experimental Section
3.1. Reagents
3.2. Collection, Extraction, and Isolation
3.2.1. Compound 9
| Position | δc | δH (Mult, J in Hz) | HMBC |
|---|---|---|---|
| 1 | 98.3 | 7.21 (1H, s) | C-9a, C-3, C-4a, C-2 |
| 2 | 147.4 | - | - |
| 3 | 140.2 | - | - |
| 4 | 134.7 | - | - |
| 4a | 144.6 | - | - |
| 5a | 146.0 | - | - |
| 6 | 144.1 | - | - |
| 7 | 113.5 | 6.82 (1H, dd, J = 7.7, 1.0) | C-5a, C-8 |
| 8 | 111.5 | 7.08 (1H, t, J = 7.7) | C-6, C-9a |
| 9 | 124.5 | 7.33 (1H, dd, J = 7.7, 1.0) | C-7, C-9a |
| 9a | 127.9 | - | - |
| 9b | 117.4 | - | - |
| 2-OMe | 57.3 | 3.94 (3H, s) | C-2 |
| 4-OMe | 61.3 | 4.15 (3H, s) | C-4 |
3.2.2. Compound 13
| Position | δc | δH (Mult, J in Hz) | HMBC |
|---|---|---|---|
| 1 | 130.6 | - | - |
| 2 | 111.4 | 6.67 (1H, d, J = 1.8) | C-3, C-4, C-6, C-1′ |
| 3 | 146.0 | - | - |
| 4 | 134.6 | - | - |
| 5 | 149.1 | - | - |
| 6 | 106.8 | 6.80 (1H, d, J = 1.8) | C-2, C-4, C-5, C-1′ |
| 1′ | 133.0 | - | - |
| 2′ | 155.4 | - | - |
| 3′ | 116.4 | 7.22 (1H, dd, J = 7.6, 1.8) | C-1′, C-2′, C-5′ |
| 4′ | 129.0 | 7.23 (1H, m) | C-2′, C-5′ |
| 5′ | 123.4 | 7.02 (1H, m) | C-1′, C-3′, C-4′, C-6′ |
| 6′ | 131.7 | 7.27 (1H, dd, J = 8.4, 1.4) | C-1, C-2′, C-4′ |
| 1′′ | 101.8 | 5.03 (1H, d, J = 7.2) | C-2′ |
| 2′′ | 75.0 | 3.43 (1H, m) | C-1′′ |
| 3′′ | 78.2 | 3.42 (1H, m) | C-4′′ |
| 4′′ | 71.3 | 3.34 (1H, m) | C-4′′ |
| 5′′ | 78.3 | 3.44 (1H, m) | C-4′′, C-3′′ |
| 6′′ | 62.5 | 3.68 (1H, dd, J = 12.0, 5.4); 3.86 (1H, dd, J = 12.0, 2.1) | C-5′′ |
| 5-OCH3 | 56.8 | 3.86 (3H, s) | C-5 |
3.3. Cell Culture
3.4. Cell Viability in Human Epidermal Melanocytes
3.5. Tyrosinase Activity in Human Epidermal Melanocytes
3.6. Melanin Contents in Human Epidermal Melanocytes
3.7. Western Blot Analysis
3.8. Real-Time PCR Analysis
3.9. Kinetic Analysis of Cellular Tyrosinase
3.10. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Videira, I.F.; Moura, D.F.; Magina, S. Mechanisms regulating melanogenesis. An. Bras. Dermatol. 2013, 88, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Plonka, P.M. Electron paramagnetic resonance as a unique tool for skin and hair research. Exp. Dermatol. 2009, 18, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; D’Ischia, M.; Misuraca, G.; Prota, G. Mechanism of inhibition of melanogenesis by hydroquinone. Biochim. Biophys. Acta 1991, 1073, 85–90. [Google Scholar] [CrossRef]
- Bilandzija, H.; Ma, L.; Parkhurst, A.; Jeffery, W.R. A potential benefit of albinism in Astyanax cavefish: Downregulation of the OCA2 gene increases tyrosine and catecholamine levels as an alternative to melanin synthesis. PLoS ONE 2013, 8, e80823. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, X.M.; Zhang, J.; Zhang, Y.Q. An efficient preparation of mulberroside a from the branch bark of mulberry and its effect on the inhibition of tyrosinase activity. PLoS ONE 2014, 9, e109396. [Google Scholar] [CrossRef] [PubMed]
- Gillbro, J.M.; Olsson, M.J. The melanogenesis and mechanisms of skin-lightening agents—Existing and new approaches. Int. J. Cosmet. Sci. 2011, 33, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Kondoh, H.; Ichihashi, M.; Hearing, V.J. Approaches to identify inhibitors of melanin biosynthesis via the quality control of tyrosinase. J. Investig. Dermatol. 2007, 127, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, T.; Yamaguchi, Y.; Batzer, J.; Coelho, S.G.; Zmudzka, B.Z.; Miller, S.A.; Wolber, R.; Beer, J.Z.; Hearing, V.J. Mechanisms of skin tanning in different racial/ethnic groups in response to ultraviolet radiation. J. Investig. Dermatol. 2005, 124, 1326–1332. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.S.; Gu, G.E.; Jo, A.R.; Bang, J.S.; Yun, H.Y.; Baek, K.J.; Kwon, N.S.; Park, K.C.; Kim, D.S. Baicalin-induced Akt activation decreases melanogenesis through downregulation of microphthalmia-associated transcription factor and tyrosinase. Eur. J. Pharmacol. 2015, 761, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Rodriguez, E.; Darias Martin, J.; Diaz Romero, C. Aloe vera as a functional ingredient in foods. Crit. Rev. Food Sci. Nutr. 2010, 50, 305–326. [Google Scholar] [CrossRef] [PubMed]
- Falodun, A.; Usifoh, C.O.; Nworgu, Z.A. Phytochemical and active column fractions of Pyrenacantha staudtii leaf extracts on isolated rat uterus. Pak. J. Pharm. Sci. 2005, 18, 31–35. [Google Scholar] [PubMed]
- Otsuka, H.; Fujioka, S.; Komiya, T.; Goto, M.; Hiramatsu, Y.; Fujimura, H. Studies on anti-inflammatory agents. V. A new anti-inflammatory constituent of Pyracantha crenulata Roem. Chem. Pharm. Bull. 1981, 29, 3099–3104. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; He, X.J.; Zhou, G.X.; Kurihara, H.; Ye, W.C.; Yao, X.S. Acylphloroglucinol glycosides from the fruits of Pyracantha fortuneana. J. Asian Nat. Prod. Res. 2008, 10, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhou, G.X.; Kurihara, H.; Ye, W.C.; Yao, X.S. Biphenyl glycosides from the fruit of Pyracantha fortuneana. J. Nat. Prod. 2006, 69, 1022–1024. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhou, G.X.; Kurihara, H.; Ye, W.C.; Yao, X.S. Fortuneanosides G–L, dibenzofuran glycosides from the fruit of Pyracantha fortuneana. Chem. Pharm. Bull. 2008, 56, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Fico, G.; Bilia, A.R.; Morelli, I.I.; Tome, F. Flavonoid distribution in Pyracantha coccinea plants at different growth phases. Biochem. Syst. Ecol. 2000, 28, 673–678. [Google Scholar] [CrossRef]
- Akar, T.; Anilan, B.; Gorgulu, A.; Akar, S.T. Assessment of cationic dye biosorption characteristics of untreated and non-conventional biomass: Pyracantha coccinea berries. J. Hazard. Mater. 2009, 168, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhou, G.X.; Kurihara, H.; Ye, W.C.; Yao, X.S. A biphenyl glycoside from Pyracantha fortuneana. Nat. Prod. Res. 2009, 23, 1163–1167. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.B.; Chang, M.J.; Wen, C.L.; Lin, Y.P.; Hsu, F.L.; Lee, M.H. Natural products of cosmetics: Analysis of extracts of plants endemic to Taiwan for the presence of tyrosinase-inhibitory, melanin-reducing, and free radical scavenging activities. J. Food Drug Anal. 2006, 14, 346–352. [Google Scholar]
- Jagan Mohan Rao, L.; Yada, H.; Ono, H.; Yoshida, M. Acylated and non-acylated flavonol monoglycosides from the Indian minor spice Nagkesar (Mammea longifolia). J. Agric. Food Chem. 2002, 50, 3143–3146. [Google Scholar] [CrossRef] [PubMed]
- Kazuma, K.; Noda, N.; Suzuki, M. Malonylated flavonol glycosides from the petals of Clitoria ternatea. Phytochemistry 2003, 62, 229–237. [Google Scholar] [CrossRef]
- Calis, I.; Kuruuzum, A.; Demirezer, L.O.; Sticher, O.; Ganci, W.; Ruedi, P. Phenylvaleric acid and flavonoid glycosides from Polygonum salicifolium. J. Nat. Prod. 1999, 62, 1101–1105. [Google Scholar] [CrossRef] [PubMed]
- Morimura, K.; Gatayama, A.; Tsukimata, R.; Matsunami, K.; Otsuka, H.; Hirata, E.; Shinzato, T.; Aramoto, M.; Takeda, Y. 5-O-glucosyldihydroflavones from the leaves of Helicia cochinchinensis. Phytochemistry 2006, 67, 2681–2685. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.L.; Chen, C.C.; Chen, Y.J.; Huang, R.L.; Shieh, B.J. Three xanthones and a benzophenone from Garcinia mangostana. J. Nat. Prod. 2001, 64, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Kokubun, T.; Harborne, J.B.; Eagles, J.; Waterman, P.G. Dibenzofuran phytoalexins from the sapwood tissue of Photinia, Pyracantha and Crataegus species. Phytochemistry 1995, 39, 1033–1037. [Google Scholar] [CrossRef]
- Yao, X.; Li, Y.; Dai, Y. Dibenzofuran Glycoside Derivatives Capable of Inhibiting Tyrosinase. Patent CN101139377A, 12 March 2008. [Google Scholar]
- Chizzali, C.; Khalil, M.N.A.; Beuerle, T.; Schuehly, W.; Richter, K.; Flachowsky, H.; Peil, A.; Hanke, M.V.; Liu, B.; Beerhues, L. Formation of biphenyl and dibenzofuran phytoalexins in the transition zones of fire blight-infected stems of Malus domestica cv. ‘Holsteiner Cox’ and Pyrus communis cv. ‘Conference’. Phytochemistry 2012, 77, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Tobin, D.J.; Shibahara, S.; Wortsman, J. Melanin pigmentation in mammalian skin and its hormonal regulation. Phys. Rev. 2004, 84, 1155–1228. [Google Scholar] [CrossRef] [PubMed]
- Negroiu, G.; Dwek, R.A.; Petrescu, S.M. Folding and maturation of tyrosinase-related protein-1 are regulated by the post-translational formation of disulfide bonds and by N-glycan processing. J. Biol. Chem. 2000, 275, 32200–32207. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, K.; Suzuki, H.; Yasumoto, K.; Tomita, Y.; Shibahara, S. Molecular cloning and functional analysis of a cDNA coding for human DOPAchrome tautomerase/tyrosinase-related protein-2. Biochim. Biophys. Acta 1994, 1217, 317–321. [Google Scholar] [CrossRef]
- Guibert, S.; Girardot, M.; Leveziel, H.; Julien, R.; Oulmouden, A. Pheomelanin coat colour dilution in French cattle breeds is not correlated with the TYR, TYRP1 and DCT transcription levels. Pigment Cell Res. 2004, 17, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Goding, C.R. Mitf from neural crest to melanoma: Signal transduction and transcription in the melanocyte lineage. Gene Dev. 2000, 14, 1712–1728. [Google Scholar] [PubMed]
- Han, E.; Chang, B.; Kim, D.; Cho, H.; Kim, S. Melanogenesis inhibitory effect of aerial part of Pueraria thunbergiana in vitro and in vivo. Arch. Dermatol. Res. 2015, 307, 57–72. [Google Scholar] [CrossRef] [PubMed]
- DiVito, K.A.; Trabosh, V.A.; Chen, Y.S.; Simbulan-Rosenthal, C.M.; Rosenthal, D.S. Inhibitor of differentiation-4 (Id4) stimulates pigmentation in melanoma leading to histiocyte infiltration. Exp. Dermatol. 2015, 24, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Bondurand, N.; Pingault, V.; Goerich, D.E.; Lemort, N.; Sock, E.; le Caignec, C.; Wegner, M.; Goossens, M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum. Mol. Genet. 2000, 9, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.J.; Ma, H.J.; Zhao, G.; Yuan, X.Y.; Zhang, P.; Liu, W.; Ma, L.J.; Lei, X.B. Additive effect of heat on the UVB-induced tyrosinase activation and melanogenesis via ERK/p38/MITF pathway in human epidermal melanocytes. Arch. Dermatol. Res. 2014, 306, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.J.; Qiu, L.; Zhou, J.J.; Guo, H.Y.; Hu, Y.H.; Li, Z.C.; Wang, Q.; Chen, Q.X.; Liu, B. Inhibitory effects of hinokitiol on tyrosinase activity and melanin biosynthesis and its antimicrobial activities. J. Enzym. Inhib. Med. Chem. 2010, 25, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.D.; Chavan, B.; Marles, L.K.; Kauser, S.; Rokos, H.; Schallreuter, K.U. A novel mechanism in control of human pigmentation by β-melanocyte-stimulating hormone and 7-tetrahydrobiopterin. J. Endocrinol. 2005, 187, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Zmijewski, M.A.; Pawelek, J. l-Tyrosine and l-dihydroxyphenylalanine as hormone-like regulators of melanocyte functions. Pigment Cell Melanoma R. 2012, 25, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Ganss, R.; Schutz, G.; Beermann, F. The mouse tyrosinase gene. Promoter modulation by positive and negative regulatory elements. J. Biol. Chem. 1994, 269, 29808–29816. [Google Scholar] [PubMed]
- Jimbow, K.; Hua, C.; Gomez, P.F.; Hirosaki, K.; Shinoda, K.; Salopek, T.G.; Matsusaka, H.; Jin, H.Y.; Yamashita, T. Intracellular vesicular trafficking of tyrosinase gene family protein in eu- and pheomelanosome biogenesis. Pigment Cell Res. 2000, 13, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Korner, A.; Pawelek, J. Activation of melanoma tyrosinase by a cyclic AMP-dependent protein kinase in a cell-free system. Nature 1977, 267, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Busca, R.; Ballotti, R. Cyclic AMP a key messenger in the regulation of skin pigmentation. Pigment. Cell Res. 2000, 13, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Yasumoto, K.; Takeda, K.; Takahashi, K.; Fukuzaki, A.; Orikasa, S.; Shibahara, S. Melanocyte-specific microphthalmia-associated transcription factor isoform activates its own gene promoter through physical interaction with lymphoid-enhancing factor 1. J. Biol. Chem. 2002, 277, 28787–28794. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Fisher, D.E. Lighting a path to pigmentation: Mechanisms of MITF induction by UV. Pigment Cell Melanoma Res. 2010, 23, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Plummer, R.S.; Shea, C.R.; Nelson, M.; Powell, S.K.; Freeman, D.M.; Dan, C.P.; Lang, D. PAX3 expression in primary melanomas and nevi. Mod. Pathol. 2008, 21, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Scholl, F.A.; Kamarashev, J.; Murmann, O.V.; Geertsen, R.; Dummer, R.; Schafer, B.W. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001, 61, 823–826. [Google Scholar] [PubMed]
- Muratovska, A.; Zhou, C.; He, S.; Goodyer, P.; Eccles, M.R. Paired-Box genes are frequently expressed in cancer and often required for cancer cell survival. Oncogene 2003, 22, 7989–7997. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, K.; Takeda, K.; Saito, H.; Watanabe, K.; Takahashi, K.; Shibahara, S. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J. 2002, 21, 2703–2714. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Singh, S.K.; Sarkar, C.; Bera, R.; Ratha, J.; Tobin, D.J.; Bhadra, R. Activation of the Mitf promoter by lipid-stimulated activation of p38-stress signalling to CREB. Pigment Cell Res. 2006, 19, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Lee, T.H.; Chan, K.J.; Hsu, F.L.; Wu, Y.C.; Lee, M.H. Anemonin is a natural bioactive compound that can regulate tyrosinase-related proteins and mRNA in human melanocytes. J. Dermatol. Sci. 2008, 49, 115–123. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, R.-D.; Chen, M.-C.; Liu, Y.-L.; Lin, Y.-T.; Lu, M.-K.; Hsu, F.-L.; Lee, M.-H. New Whitening Constituents from Taiwan-Native Pyracantha koidzumii: Structures and Tyrosinase Inhibitory Analysis in Human Epidermal Melanocytes. Int. J. Mol. Sci. 2015, 16, 28598-28613. https://doi.org/10.3390/ijms161226115
Lin R-D, Chen M-C, Liu Y-L, Lin Y-T, Lu M-K, Hsu F-L, Lee M-H. New Whitening Constituents from Taiwan-Native Pyracantha koidzumii: Structures and Tyrosinase Inhibitory Analysis in Human Epidermal Melanocytes. International Journal of Molecular Sciences. 2015; 16(12):28598-28613. https://doi.org/10.3390/ijms161226115
Chicago/Turabian StyleLin, Rong-Dih, Mei-Chuan Chen, Yan-Ling Liu, Yi-Tzu Lin, Mei-Kuang Lu, Feng-Lin Hsu, and Mei-Hsien Lee. 2015. "New Whitening Constituents from Taiwan-Native Pyracantha koidzumii: Structures and Tyrosinase Inhibitory Analysis in Human Epidermal Melanocytes" International Journal of Molecular Sciences 16, no. 12: 28598-28613. https://doi.org/10.3390/ijms161226115