A Transcriptomic Study of Maternal Thyroid Adaptation to Pregnancy in Rats
Abstract
:1. Introduction
2. Results
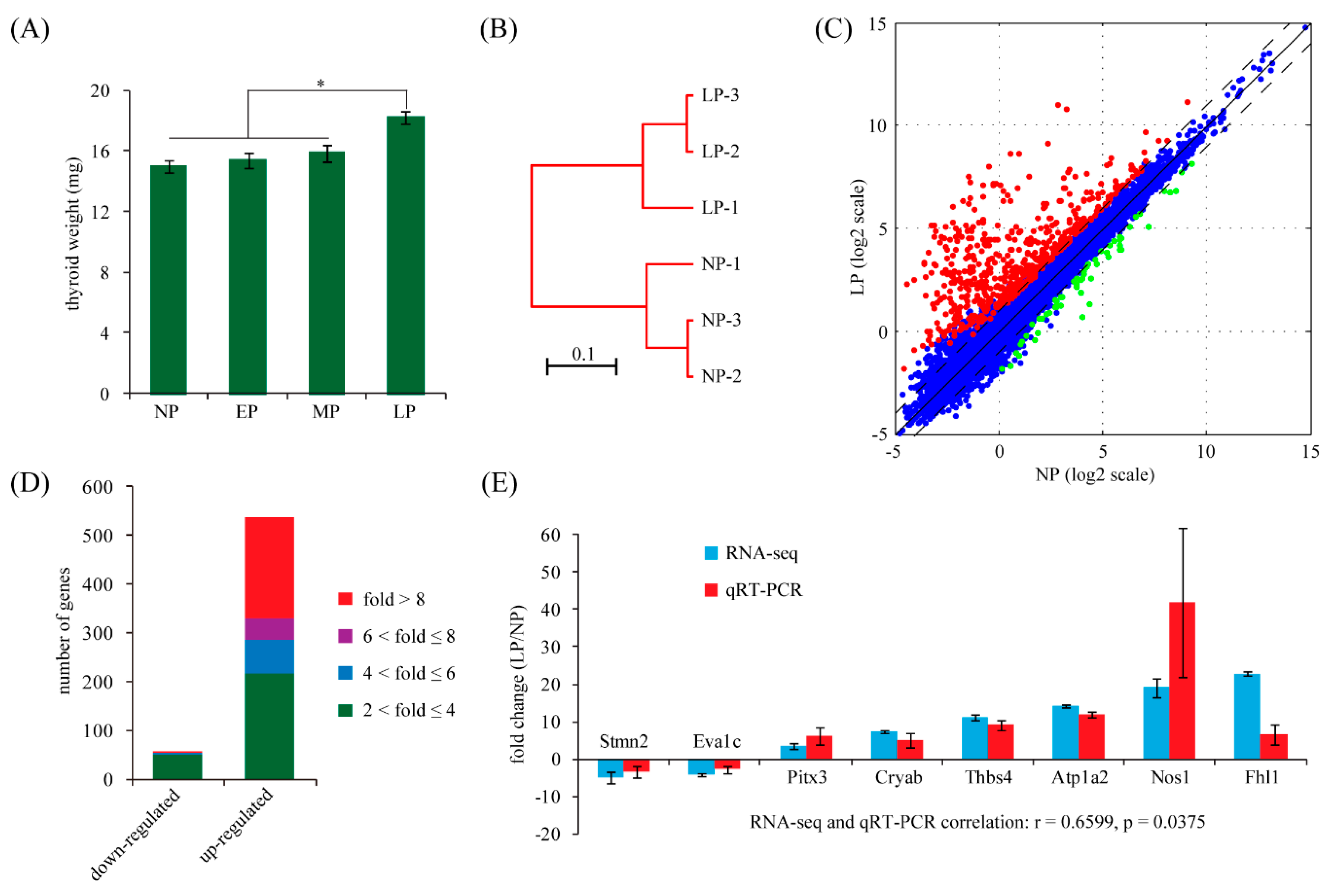
2.1. Maternal Thyroid Weight Changes during Pregnancy in Rats
2.2. Identification of Differentially Expressed Genes in Maternal Thyroid in Late Pregnancy
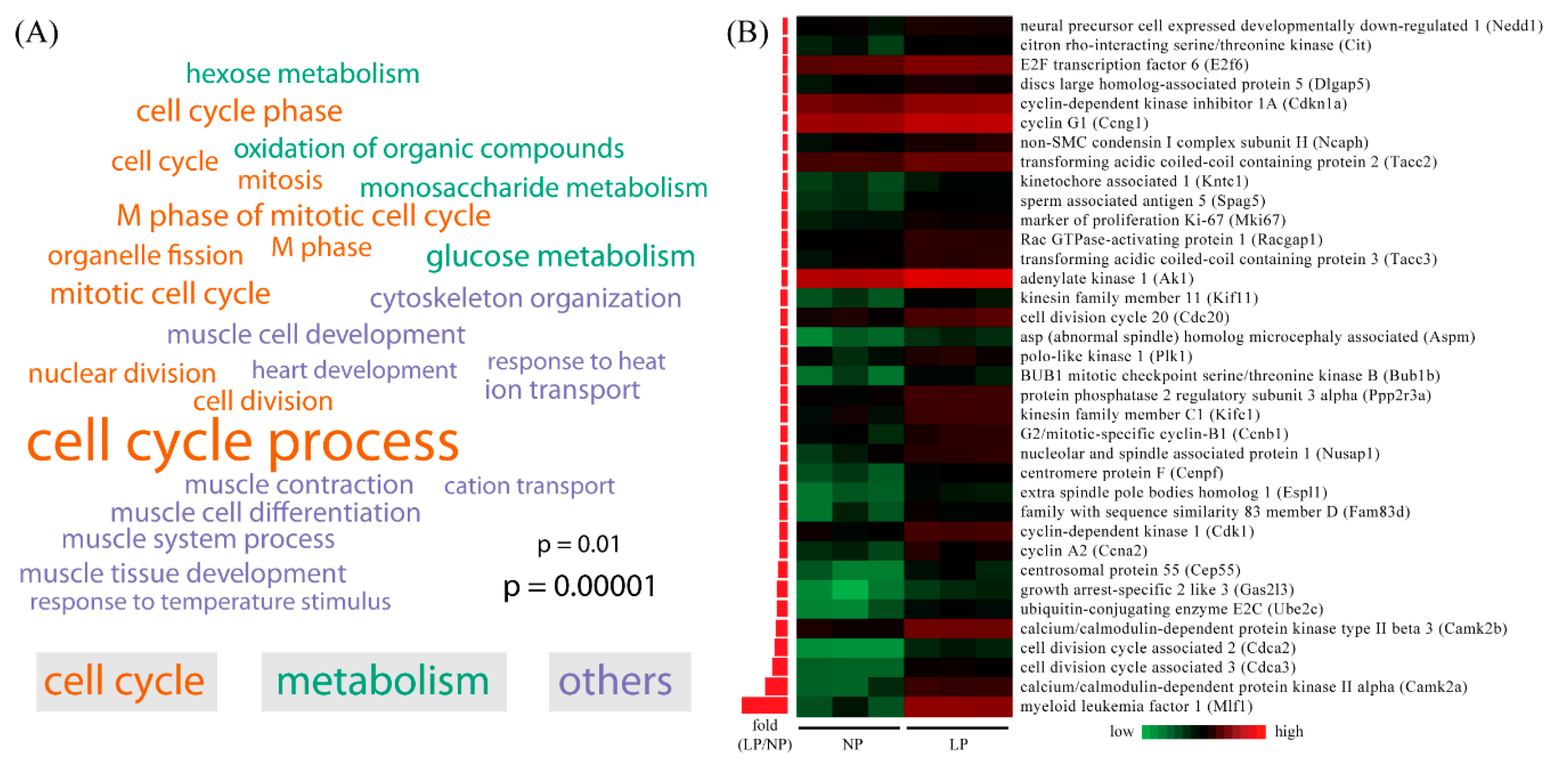
2.3. Gene Ontology (GO) Analysis of Differentially Expressed Genes in Maternal Thyroid in Late Pregnancy
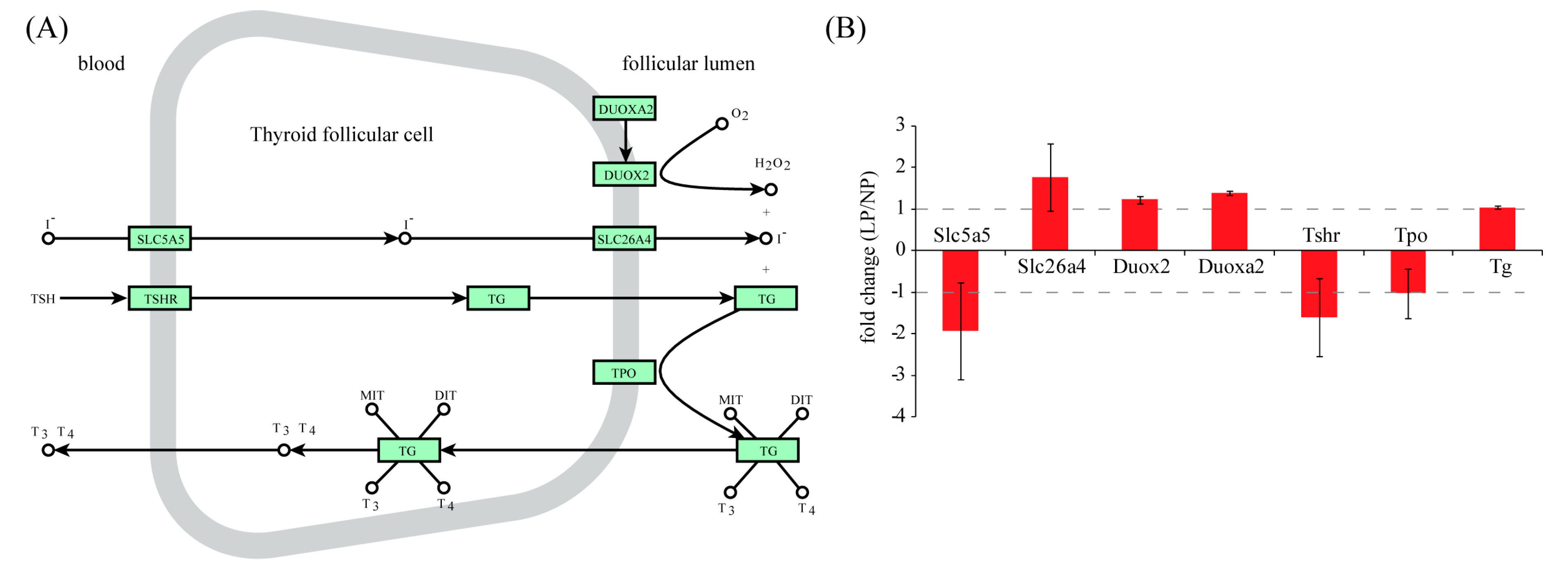
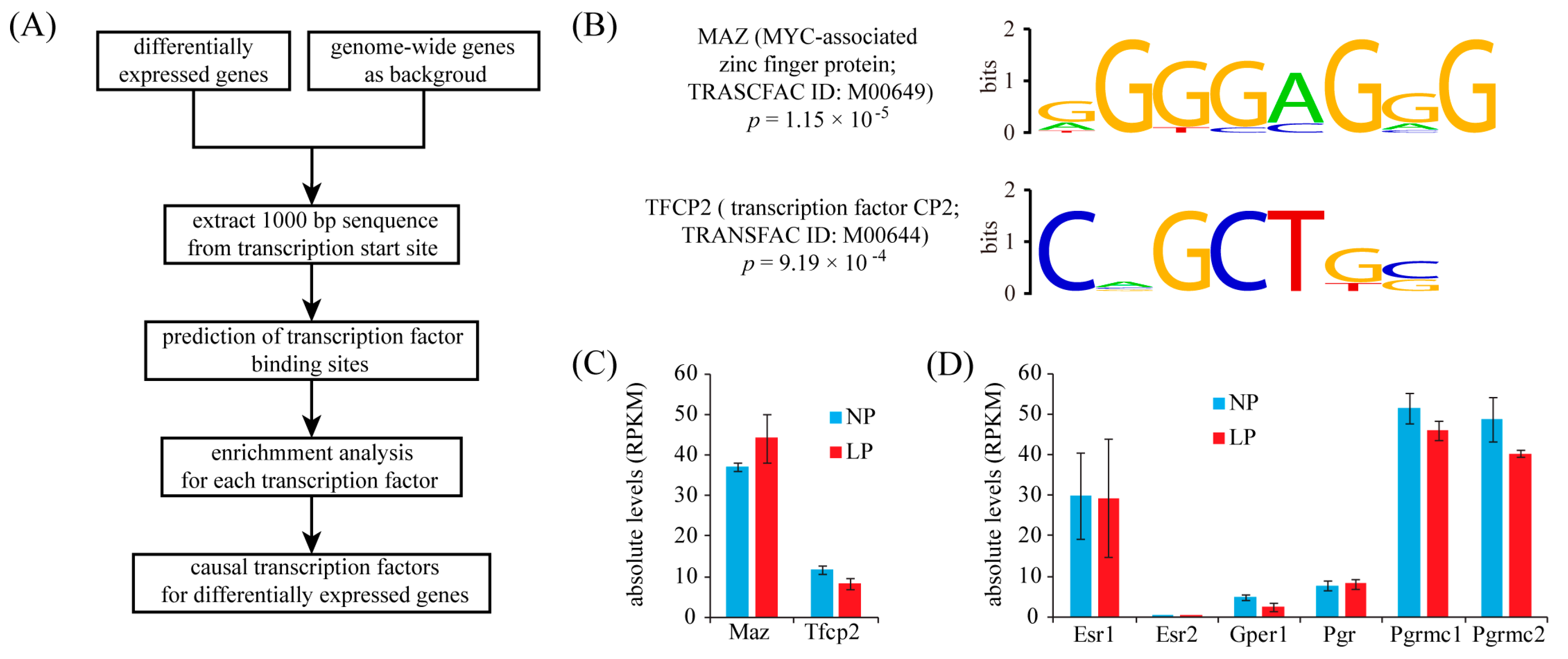
2.4. Identification of Transcription Factor Binding Sites (TFBS) in the Promoters of Up-Regulated Genes
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. RNA-Seq Analysis
4.3. Validation by Quantitative RT-PCR
4.4. Functional Clustering Analysis
4.5. Analysis of Transcription Factor Binding Sites (TFBS)
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bustamante, J.J.; Copple, B.L.; Soares, M.J.; Dai, G. Gene profiling of maternal hepatic adaptations to pregnancy. Liver Int. 2010, 30, 406–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacovetti, C.; Abderrahmani, A.; Parnaud, G.; Jonas, J.C.; Peyot, M.L.; Cornu, M.; Laybutt, R.; Meugnier, E.; Rome, S.; Thorens, B.; et al. Micrornas contribute to compensatory beta cell expansion during pregnancy and obesity. J. Clin. Investig. 2012, 122, 3541–3551. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.; Leinwand, L.A. Pregnancy as a cardiac stress model. Cardiovasc. Res. 2014, 101, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.H.; Ertelt, J.M.; Xin, L.; Way, S.S. Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature 2012, 490, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Feely, J. The physiology of thyroid function in pregnancy. Postgrad. Med. J. 1979, 55, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Berghout, A.; Wiersinga, W. Thyroid size and thyroid function during pregnancy: An analysis. Eur. J. Endocrinol. 1998, 138, 536–542. [Google Scholar] [CrossRef]
- Krassas, G.E.; Poppe, K.; Glinoer, D. Thyroid function and human reproductive health. Endocr. Rev. 2010, 31, 702–755. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, N.; Nishikawa, M.; Horimoto, M.; Yoshimura, M.; Sawaragi, S.; Horikoshi, Y.; Sawaragi, I.; Inada, M. Thyroid-stimulating activity in sera of normal pregnant women. J. Clin. Endocrinol. Metab. 1989, 69, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Pekonen, F.; Alfthan, H.; Stenman, U.H.; Ylikorkala, O. Human chorionic gonadotropin (hcg) and thyroid function in early human pregnancy: Circadian variation and evidence for intrinsic thyrotropic activity of hcg. J. Clin. Endocrinol. Metab. 1988, 66, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Amino, N.; Tamaki, H.; Mitsuda, N.; Miyai, K.; Tanizawa, O. Physiologic thyroid activation in normal early pregnancy is induced by circulating hcg. Obstet. Gynecol. 1990, 75, 775–778. [Google Scholar] [PubMed]
- Allan, W.C.; Haddow, J.E.; Palomaki, G.E.; Williams, J.R.; Mitchell, M.L.; Hermos, R.J.; Faix, J.D.; Klein, R.Z. Maternal thyroid deficiency and pregnancy complications: Implications for population screening. J. Med. Screen. 2000, 7, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.Z.; Haddow, J.E.; Faix, J.D.; Brown, R.S.; Hermos, R.J.; Pulkkinen, A.; Mitchell, M.L. Prevalence of thyroid deficiency in pregnant women. Clin. Endocrinol. 1991, 35, 41–46. [Google Scholar] [CrossRef]
- Stagnaro-Green, A. Overt hyperthyroidism and hypothyroidism during pregnancy. Clin. Obstet. Gynecol. 2011, 54, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Stagnaro-Green, A. Approach to the patient with postpartum thyroiditis. J. Clin. Endocrinol. Metab. 2012, 97, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, M.A.; Glidewell-Kenney, C.; Jimenez, M.A.; Ahearn, P.C.; Weiss, J.; Jameson, J.L.; Levine, J.E. New insights into the classical and non-classical actions of estrogen: Evidence from estrogen receptor knock-out and knock-in mice. Mol. Cell. Endocrinol. 2008, 290, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Large, M.J.; DeMayo, F.J. The regulation of embryo implantation and endometrial decidualization by progesterone receptor signaling. Mol. Cell. Endocrinol. 2012, 358, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Prossnitz, E.R.; Barton, M. Estrogen biology: New insights into gper function and clinical opportunities. Mol. Cell. Endocrinol. 2014, 389, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Gellersen, B.; Fernandes, M.S.; Brosens, J.J. Non-genomic progesterone actions in female reproduction. Hum. Reprod. Updat. 2009, 15, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, C.; Ito, M.; Yamada, H.; Yamakawa, N.; Yoshida, H.; Date, A.; Watanabe, M.; Hidaka, Y.; Iwatani, Y.; Miyauchi, A.; et al. Genes that characterize T3-predominant graves’ thyroid tissues. Eur. J. Endocrinol. 2013, 168, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.H.; Wu, J.C.; Chang, K.J.; Liaw, K.Y.; Wang, S.M. Distribution of the cadherin-catenin complex in normal human thyroid epithelium and a thyroid carcinoma cell line. J. Cell. Biochem. 1998, 70, 330–337. [Google Scholar] [CrossRef]
- Baris, O.; Mirebeau-Prunier, D.; Savagner, F.; Rodien, P.; Ballester, B.; Loriod, B.; Granjeaud, S.; Guyetant, S.; Franc, B.; Houlgatte, R.; et al. Gene profiling reveals specific oncogenic mechanisms and signaling pathways in oncocytic and papillary thyroid carcinoma. Oncogene 2005, 24, 4155–4161. [Google Scholar] [CrossRef] [PubMed]
- Galton, V.A. Thyroxine metabolism and thyroid function in the pregnant rat. Endocrinology 1968, 82, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Calvo, R.; Obregon, M.J.; Ruiz de Ona, C.; Ferreiro, B.; Escobar Del Rey, E.; Morreale de Escobar, G. Thyroid hormone economy in pregnant rats near term: A “physiological” animal model of nonthyroidal illness? Endocrinology 1990, 127, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Candille, S.I.; Choi, Y.; Xie, D.; Jiang, L.; Li-Pook-Than, J.; Tang, H.; Snyder, M. Variation and genetic control of protein abundance in humans. Nature 2013, 499, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Bossone, S.A.; Asselin, C.; Patel, A.J.; Marcu, K.B. Maz, a zinc finger protein, binds to c-myc and c2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. USA 1992, 89, 7452–7456. [Google Scholar] [CrossRef] [PubMed]
- Pyrc, J.J.; Moberg, K.H.; Hall, D.J. Isolation of a novel cDNA encoding a zinc-finger protein that binds to two sites within the c-myc promoter. Biochemistry 1992, 31, 4102–4110. [Google Scholar] [CrossRef] [PubMed]
- Amati, B.; Land, H. Myc-max-mad: A transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 1994, 4, 102–108. [Google Scholar] [CrossRef]
- Hsu, S.F.; Goan, Y.G.; Tsai, H.Y.; Lin, Y.R.; Liu, R.T.; Cheng, J.T. An upstream regulatory element confers orientation-independent enhancement of the TSG101 promoter activity in transformed cells. Mol. Biol. Rep. 2012, 39, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Veljkovic, J.; Hansen, U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene 2004, 343, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. Tophat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. Htseq—A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2015. Nucleic Acids Res. 2015, 43, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. David bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Schug, J. Using tess to predict transcription factor binding sites in DNA sequence. Curr. Protoc. Bioinform. 2008. [Google Scholar] [CrossRef]
- Wingender, E.; Dietze, P.; Karas, H.; Knuppel, R. Transfac: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996, 24, 238–241. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.-L.; Wang, T.-S.; Zhao, M.; Peng, Y.; Fu, Y.-S. A Transcriptomic Study of Maternal Thyroid Adaptation to Pregnancy in Rats. Int. J. Mol. Sci. 2015, 16, 27339-27349. https://doi.org/10.3390/ijms161126030
Liu J-L, Wang T-S, Zhao M, Peng Y, Fu Y-S. A Transcriptomic Study of Maternal Thyroid Adaptation to Pregnancy in Rats. International Journal of Molecular Sciences. 2015; 16(11):27339-27349. https://doi.org/10.3390/ijms161126030
Chicago/Turabian StyleLiu, Ji-Long, Tong-Song Wang, Miao Zhao, Ying Peng, and Yong-Sheng Fu. 2015. "A Transcriptomic Study of Maternal Thyroid Adaptation to Pregnancy in Rats" International Journal of Molecular Sciences 16, no. 11: 27339-27349. https://doi.org/10.3390/ijms161126030





