Antibacterial Activity of Shikimic Acid from Pine Needles of Cedrus deodara against Staphylococcus aureus through Damage to Cell Membrane
Abstract
:1. Introduction
2. Results and Discussion
2.1. Antibacterial Activity of Shikimic Acid (SA) to Staphylococcus aureus (S. aureus)
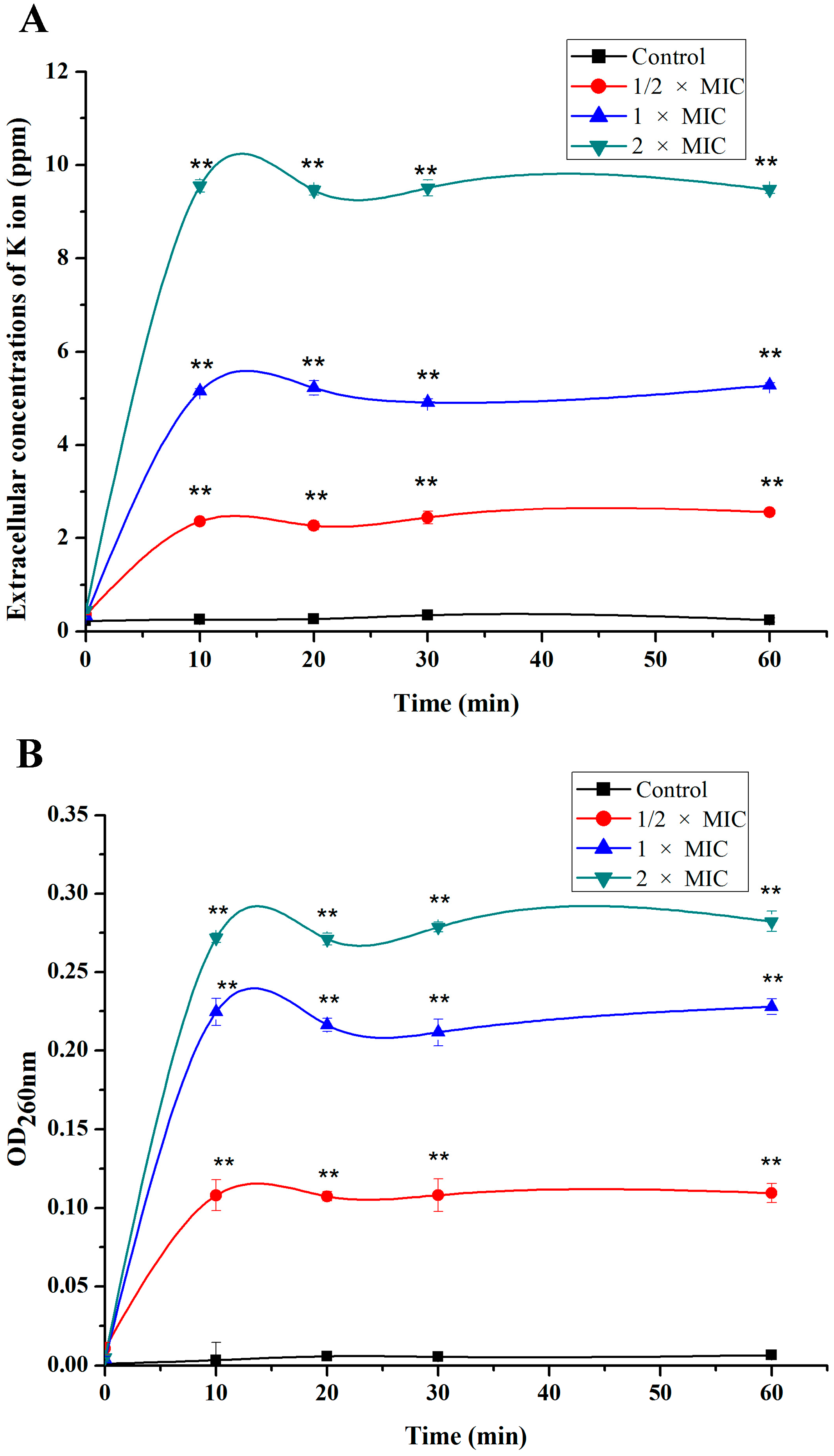
2.2. Leakage of Cell Constituents Induced by SA
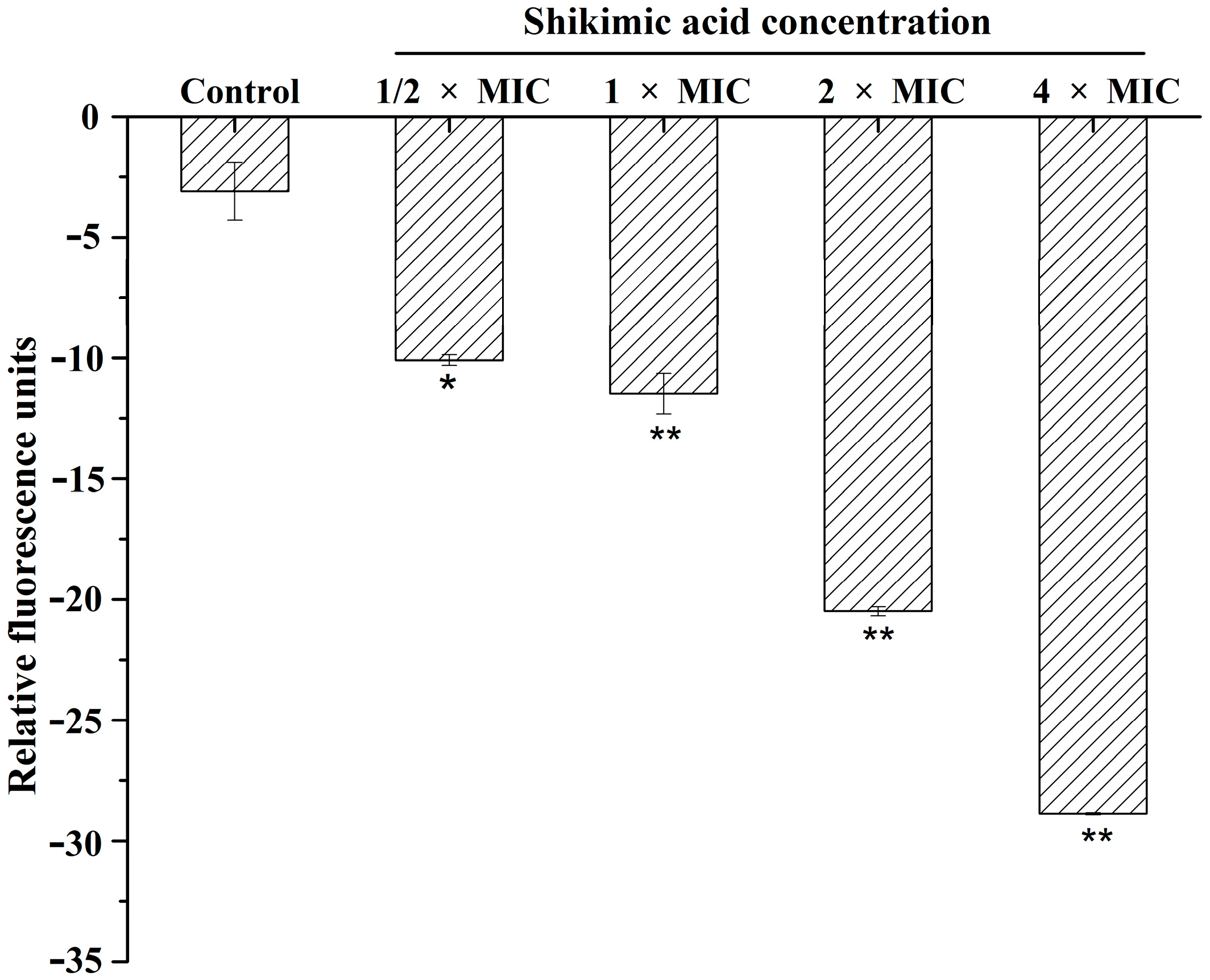
2.3. Effect of SA on Membrane Potential
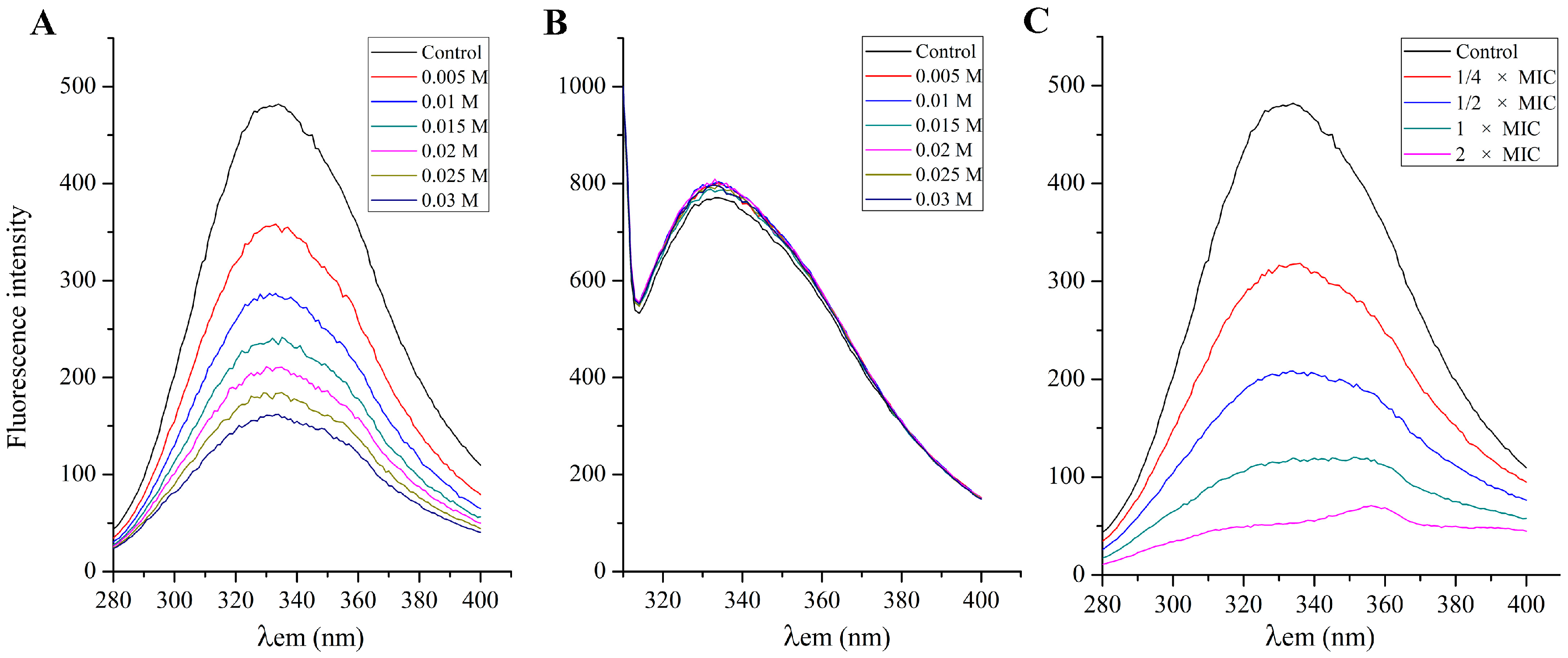
2.4. Membrane Integrity
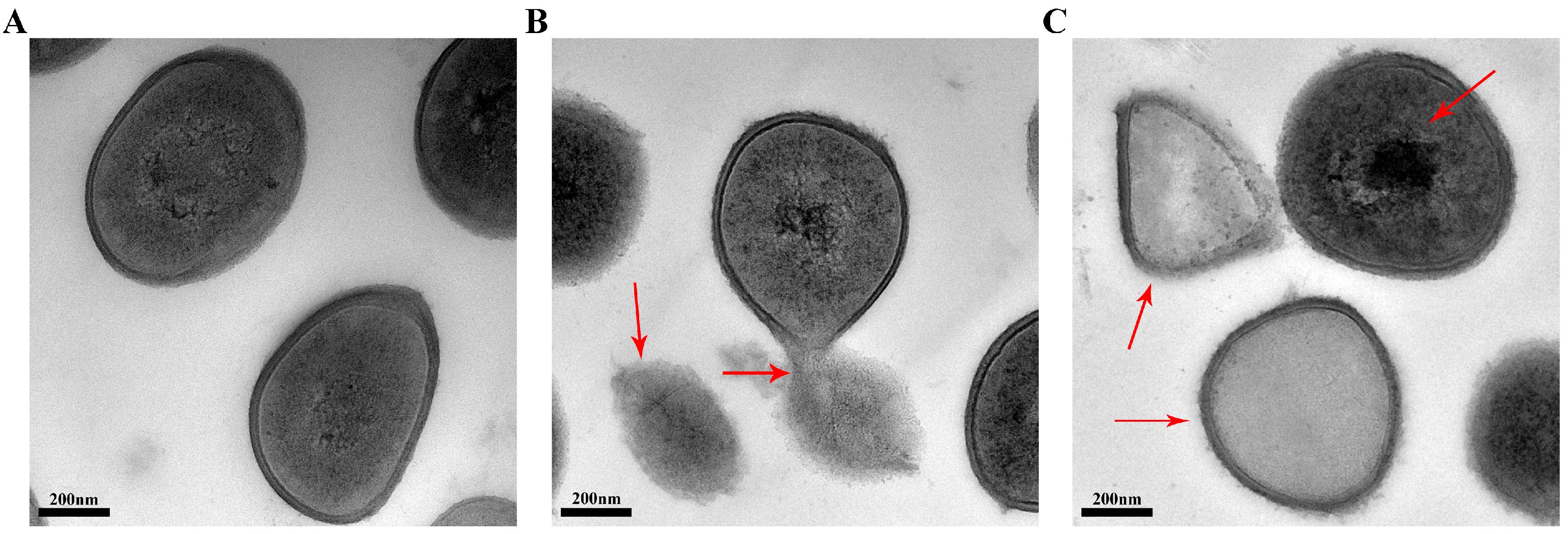
2.5. Transmission Electron Microscopy (TEM)

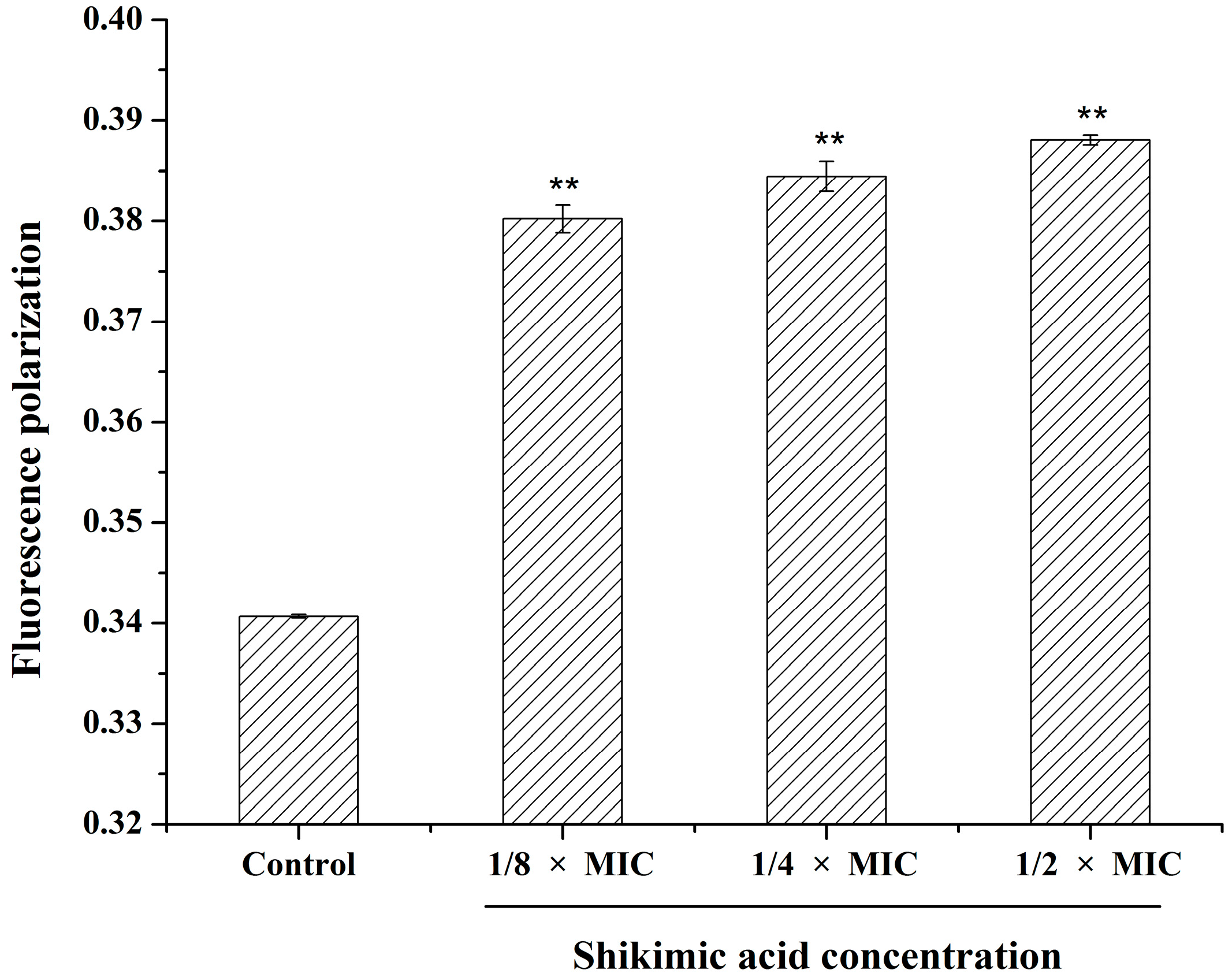
2.6. Effect of SA on the S. aureus Cell Membrane Fluidity
2.7. Effect of SA on the S. aureus Cell Membrane Protein
3. Materials and Methods
3.1. Materials
3.2. Assay for Antibacterial Activity
3.3. Measurement of the Potassium Efflux
3.4. Measurement of the Nucleotide Leakage
3.5. Membrane Potential Assay
3.6. Membrane Integrity
3.7. TEM
3.8. Membrane Fluidity Assay
3.9. Effect of SA on S. aureus Membrane Protein
3.10. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Le-Loir, Y.; Baron, F.; Gautier, M. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2003, 2, 63–76. [Google Scholar] [PubMed]
- Braden, C.R.; Tauxe, R.V. Emerging trends in foodborne diseases. Infect. Dis. Clin. N. Am. 2013, 27, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Elaine, S.; Robert, M.H.; Frederick, J.A.; Robert, V.T.; Marc-Alain, W.; Sharon, L.R.; Jeffery, L.J.; Patricia, M.G. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. J. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Kadariya, J.; Smith, T.C.; Thapaliya, D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Rozemeijer, W.; Fink, P.; Rojas, E.; Jones, C.H.; Pavliakova, D.; Giardina, P.; Murphy, E.; Liberator, P.; Jiang, Q.; Girgenti, D.; et al. Evaluation of approaches to monitor Staphylococcus aureus virulence factor expression during human disease. PLoS ONE 2015, 10, e0116945. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, J.E.; Muthaiyan, A.; Dupre, J.M.; Ricke, S.C. Staphylococcus aureus and understanding the factors that impact enterotoxin production in foods: A review. Food Control 2014, 2015, 1–14. [Google Scholar] [CrossRef]
- Michailidis, P.A.; Krokida, M.K. Drying and dehydration processes in food preservation and processing. In Conventional and Advanced Food Processing Technologies; Bhattacharya, S., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; pp. 1–32. [Google Scholar]
- Calo, J.R.; Crandall, P.G.; O’Bryan, C.A.; Ricke, S.C. Essential oils as antimicrobials in food systems—A review. Food Control 2015, 54, 111–119. [Google Scholar] [CrossRef]
- Tripathi, P.; Rawat, G.; Yadav, S.; Saxena, R.K. Shikimic acid, a base compound for the formulation of swine/avian flu drug: Statistical optimization, fed-batch and scale up studies along with its application as an antibacterial agent. Antonie Van Leeuwenhoek 2015, 107, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Just, J.; Deans, B.J.; Olivier, W.J.; Paull, B.; Bissember, A.C.; Smith, J.A. New method for the rapid extraction of natural products: Efficient isolation of shikimic acid from star anise. Org. Lett. 2015, 17, 2428–2430. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hou, K.; Li, S.; Zu, Y.; Yang, L. Extraction and chromatographic determination of shikimic acid in chinese conifer needles with 1-benzyl-3-methylimidazolium bromide ionic liquid aqueous solutions. J. Anal. Methods Chem. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Rabelo, T.; Zeidán-Chuliá, F.; Caregnato, F.; Schnorr, C.; Gasparotto, J.; Serafini, M.; de Souza-Araújo, A.; Quintans-Junior, L.; Moreira, J.; Gelain, D. In vitro neuroprotective effect of shikimic acid against hydrogen peroxide-induced oxidative stress. J. Mol. Neurosci. 2015, 56, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Xiang, H.; Sun, Y.; Qiu, L.; Chen, D.; Deng, C.; Chen, W. Monopalmityloxy shikimic acid: Enzymatic synthesis and anticoagulation activity evaluation. Appl. Biochem. Biotechnol. 2009, 158, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Sun, J.; You, H.; Lv, J.; Sun, J.; Dong, Y. Anti-inflammatory effect of 3,4-oxo-isopropylidene-shikimic acid on acetic acid-induced colitis in rats. Inflammation 2012, 35, 1872–1879. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.C.; He, Q.; Sun, Q.; Zhong, K.; Gao, H. Antibacterial activity of water-soluble extract from pine needles of Cedrus deodara. Int. J. Food Microbiol. 2012, 153, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Sila, A.; Hedhili, K.; Przybylski, R.; Ellouz-Chaabouni, S.; Dhulster, P.; Bougatef, A.; Nedjar-Arroume, N. Antibacterial activity of new peptides from barbel protein hydrolysates and mode of action via a membrane damage mechanism against Listeria monocytogenes. J. Funct. Foods 2014, 11, 322–329. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Rao, S.; Sun, J.; Ma, C.; Li, J. p-Coumaric acid kills bacteria through dual damage mechanisms. Food Control 2012, 25, 550–554. [Google Scholar] [CrossRef]
- Lacombe, A.; McGivney, C.; Tadepalli, S.; Sun, X.; Wu, V.C. The effect of American cranberry (Vaccinium macrocarpon) constituents on the growth inhibition, membrane integrity, and injury of Escherichia coli O157:H7 and Listeria monocytogenes in comparison to Lactobacillus rhamnosus. Food Microbiol. 2013, 34, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wang, X.; Xu, Y.; Zhang, B.; Xia, X. Antimicrobial effect and mode of action of chlorogenic acid on Staphylococcus aureus. Eur. Food Res. Technol. 2014, 238, 589–596. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Zhu, S.; Ma, C.; Wang, Z. Antibacterial activity and mechanism of action of chlorogenic acid. J. Food Sci. 2011, 76, M398–M403. [Google Scholar] [CrossRef] [PubMed]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.-U.; Egli, T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD baclight kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. [Google Scholar] [CrossRef] [PubMed]
- Stocks, S.M. Mechanism and use of the commercially available viability stain, BacLight. Cytom. Part A 2004, 61A, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Otto, C.C.; Cunningham, T.M.; Hansen, M.R.; Haydel, S.E. Effects of antibacterial mineral leachates on the cellular ultrastructure, morphology, and membrane integrity of Escherichia coli and methicillin-resistant Staphylococcus aureus. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Ulrih, N.P.; Maričić, M.; Ota, A.; Šentjurc, M.; Abram, V. Kaempferol and quercetin interactions with model lipid membranes. Food Res. Int. 2015, 71, 146–154. [Google Scholar] [CrossRef]
- Vincent, M.; England, L.S.; Trevors, J.T. Cytoplasmic membrane polarization in Gram-positive and Gram-negative bacteria grown in the absence and presence of tetracycline. Biochim. Biophys. Acta Gen. Subj. 2004, 1672, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Huang, X.; Xu, S.; Ma, F. The antibacterial activity of ceramsite coated by silver nanoparticles in micropore. Appl. Biochem. Biotechnol. 2015, 176, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Li, X.; Yuan, L.; Ge, L.; Zhang, B.; Zhou, S. Interaction of houttuyfonate homologues with the cell membrane of Gram-positive and Gram-negative bacteria. Colloids Surf. Physicochem. Eng. Asp. 2007, 301, 412–418. [Google Scholar] [CrossRef]
- Xing, K.; Chen, X.G.; Liu, C.S.; Cha, D.S.; Park, H.J. Oleoyl-chitosan nanoparticles inhibits Escherichia coli and Staphylococcus aureus by damaging the cell membrane and putative binding to extracellular or intracellular targets. Int. J. Food Microbiol. 2009, 132, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Shi, Y.H.; Tang, Y.L.; Le, G.W. The membrane action mechanism of analogs of the antimicrobial peptide Buforin 2. Peptides 2009, 30, 1421–1427. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, W.; Wang, T.; Li, N.; Mu, H.; Zhang, J.; Duan, J. Unveiling the mode of action of two antibacterial tanshinone derivatives. Int. J. Mol. Sci. 2015, 16, 17668–17681. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, E.; García, S.; Heredia, N. Extracts of edible and medicinal plants damage membranes of Vibrio cholerae. Appl. Environ. Microbiol. 2010, 76, 6888–6894. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Lu, Z.; Zhang, X.; Li, C.; Jia, Y. Polymer-Ag nanocomposites with enhanced antimicrobial activity against bacterial infection. ACS Appl. Mater. Interfaces 2014, 6, 15813–15821. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, J.; Wu, Y.; Liu, X.; Zhong, K.; Huang, Y.; Gao, H. Antibacterial Activity of Shikimic Acid from Pine Needles of Cedrus deodara against Staphylococcus aureus through Damage to Cell Membrane. Int. J. Mol. Sci. 2015, 16, 27145-27155. https://doi.org/10.3390/ijms161126015
Bai J, Wu Y, Liu X, Zhong K, Huang Y, Gao H. Antibacterial Activity of Shikimic Acid from Pine Needles of Cedrus deodara against Staphylococcus aureus through Damage to Cell Membrane. International Journal of Molecular Sciences. 2015; 16(11):27145-27155. https://doi.org/10.3390/ijms161126015
Chicago/Turabian StyleBai, Jinrong, Yanping Wu, Xiaoyan Liu, Kai Zhong, Yina Huang, and Hong Gao. 2015. "Antibacterial Activity of Shikimic Acid from Pine Needles of Cedrus deodara against Staphylococcus aureus through Damage to Cell Membrane" International Journal of Molecular Sciences 16, no. 11: 27145-27155. https://doi.org/10.3390/ijms161126015




