Long-Term Anti-Allodynic Effect of Immediate Pulsed Radiofrequency Modulation through Down-Regulation of Insulin-Like Growth Factor 2 in a Neuropathic Pain Model
Abstract
:1. Introduction
2. Results
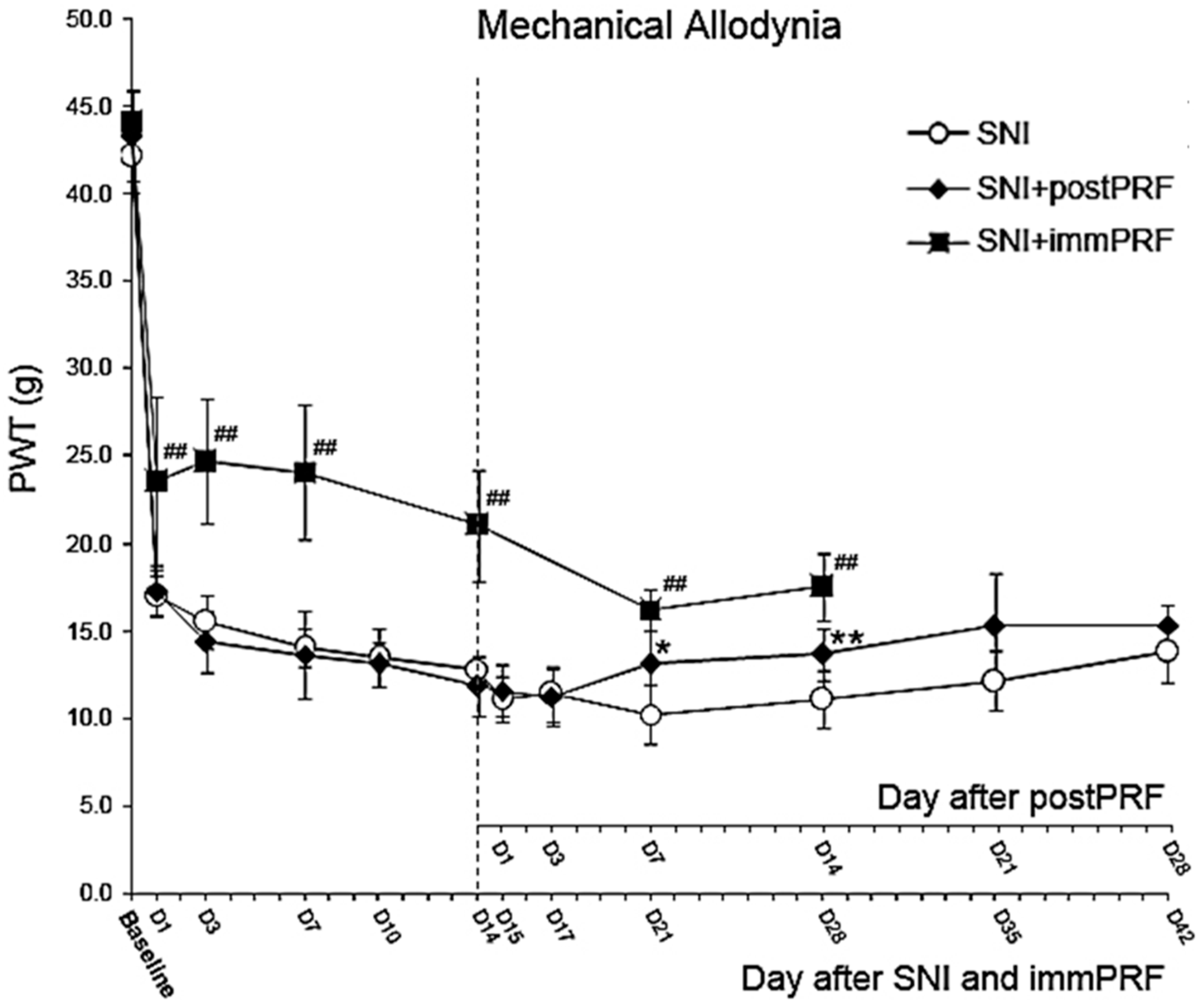
2.1. Anti-Allodynic Effect of Immediate PRF Treatment
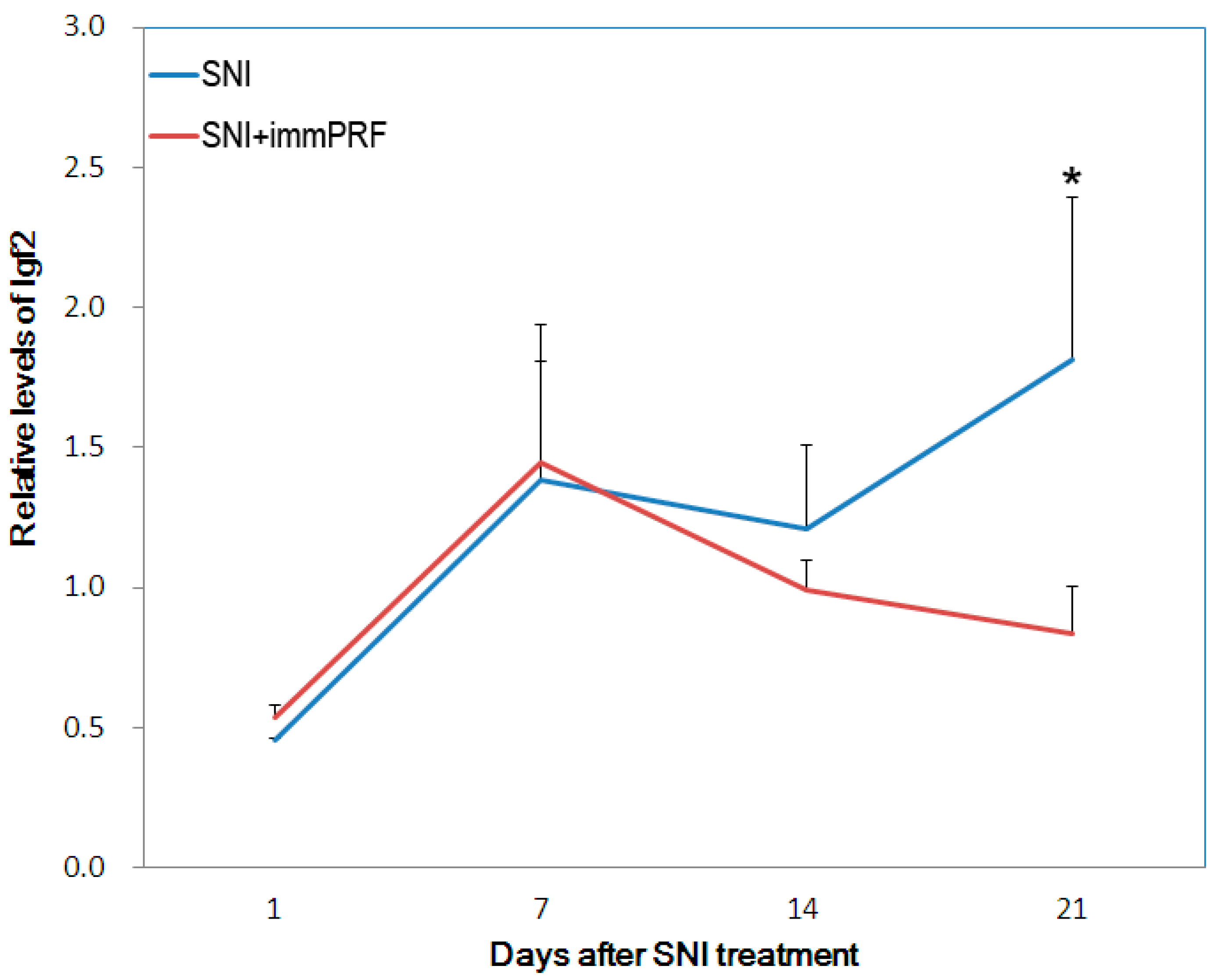
2.2. Bioinformatic Analyses from Oligonucleotide Microarray Hybridization and Gene Validation

| GO Term | GO ID | Subontology | Genes | Pop. % | Study % | adj. P |
|---|---|---|---|---|---|---|
| extracellular matrix | GO:0031012 | Cellular Component | Bgn, Cilp, Col1a1, Col1a2, Col3a1, Col4a5, Cpxm2, Ctsg, Dcn, Lox, Loxl1, Ltbp1, Mfap5, Mgp, Pcolce, Rpesp, Serpinf1, Smoc2, Vwf | 2.18% | 28.36% | <0.001 |
| extracellular region part | GO:0044421 | Cellular Component | Anxa1, Apoc3, Bgn, Ccl19, Cilp, Col1a1, Col1a2, Col3a1, Col4a5, Cpxm2, Ctsg, Cxcl9, Dcn, Igf2, Igfbp3, Igfbp4, Lox, Ltbp1, Mfap5, Mgp, Pcolce, Serpinf1, Smoc2, Sostdc1, Vwf | 5.36% | 37.31% | <0.001 |
| extracellular region | GO:0005576 | Cellular Component | Anxa1, Apoc3, Apol3, Bgn, Ccl19, Cilp, Col1a1, Col1a2, Col3a1, Col4a5, Cpxm2, Ctsg, Cxcl9, Dcn, Igf2, Igfbp3, Igfbp4, Lox, Ltbp1, Mfap5, Mgp, Olfml1, Otos, Pcolce, Serpinf1, Sfrp5, Smoc2, Sostdc1, Vwf | 8.48% | 44.78% | <0.001 |
| extracellular matrix part | GO:0044420 | Cellular Component | Cilp, Col1a1, Col1a2, Col3a1, Col4a5, Dcn, Lox, Mfap5, Smoc2 | 0.90% | 13.43% | <0.001 |
2.3. Determination of ERK1/2 Phosphorylation in the Presence of IGF2

3. Discussion
3.1. Ckm, RGD1561759, and IGF2 Decreased in Spared-Nerve-Injured Rats with immPRF Treatment
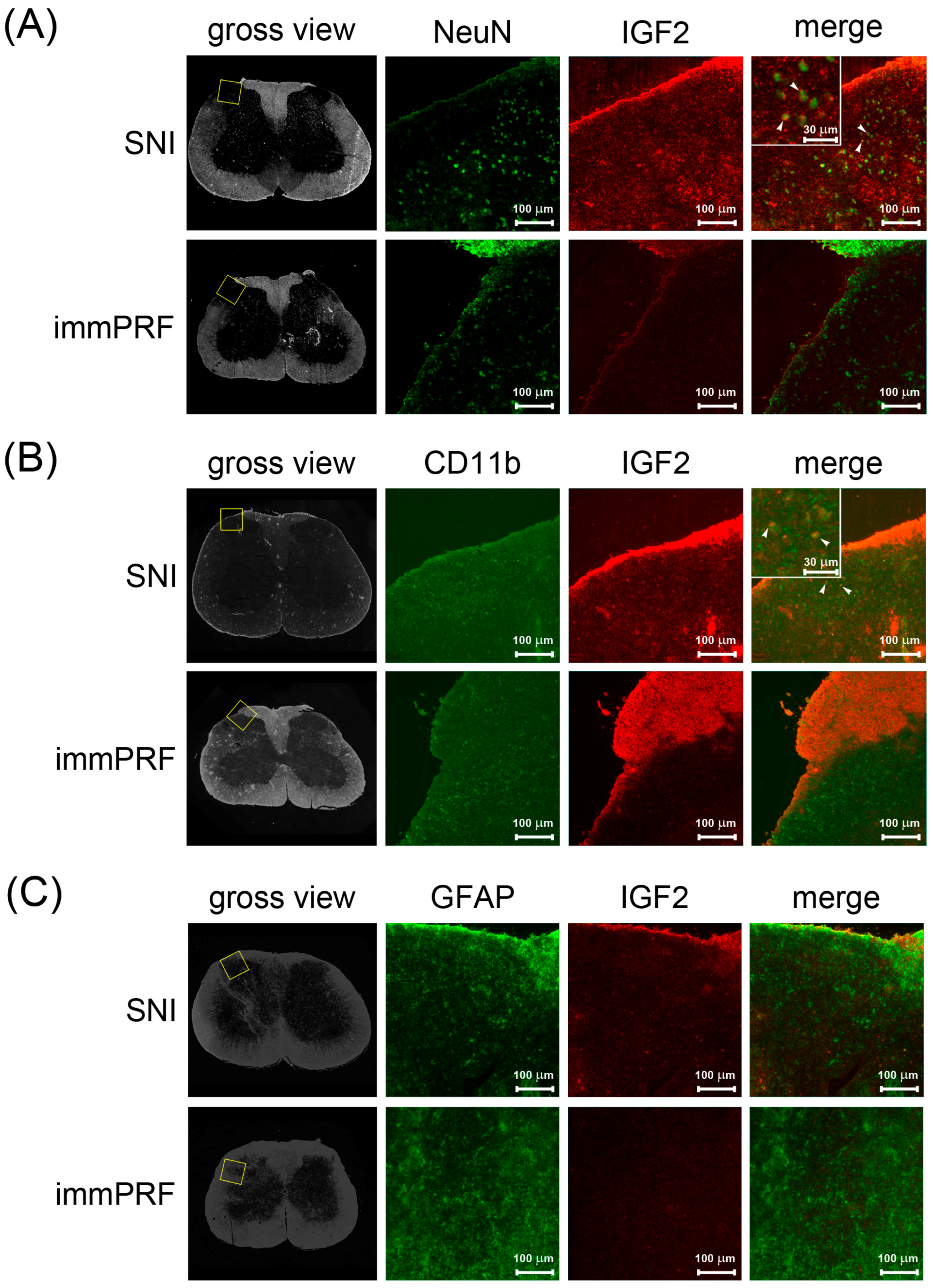
3.2. Human IGF2 Induced ERK1/2 Phosphorylation during SNI-Induced Neuropathic Pain and Showed Predominantly in Microglial Cells
3.3. Down-Regulation of IGF2 May Be the Result of Immediate PRF
4. Materials and Methods
4.1. Rats, Neuropathic Pain Model
4.2. Total RNA Extraction and Microarray Hybridization
4.3. Bioinformatics Analysis of Rat Whole-Genome Microarrays
4.4. Reverse Transcription and Validation of Rat IGF2 Expression
4.5. Glioblastoma Cells and Neuroblastoma Cells
4.6. Immunofluorescent Staining of IGF2, Phosphorylated ERK, Glial Fibrillary Acidic Protein (GFAP), NeuN, and CD11b in the Dorsal Horn Regions of Rats
4.7. Western Blotting of IGF2-Dependent Phosphorylated ERK in Human Neuronal Cells
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Byrd, D.; Mackey, S. Pulsed radiofrequency for chronic pain. Curr. Pain Headache Rep. 2008, 12, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Van Boxem, K.; van Eerd, M.; Brinkhuizen, T.; Patijn, J.; van Kleef, M.; van Zundert, J. Radiofrequency and pulsed radiofrequency treatment of chronic pain syndromes: The available evidence. Pain Pract. 2008, 8, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Chua, N.H.; Vissers, K.C.; Sluijter, M.E. Pulsed radiofrequency treatment in interventional pain management: Mechanisms and potential indications—A review. Acta Neurochir. 2011, 153, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, T.T.; Kraemer, J.; Nagda, J.V.; Aner, M.; Bajwa, Z.H. Response to pulsed and continuous radiofrequency lesioning of the dorsal root ganglion and segmental nerves in patients with chronic lumbar radicular pain. Pain Phys. 2008, 11, 137–144. [Google Scholar]
- Van Zundert, J.; Patijn, J.; Kessels, A.; Lame, I.; van Suijlekom, H.; van Kleef, M. Pulsed radiofrequency adjacent to the cervical dorsal root ganglion in chronic cervical radicular pain: A double blind sham controlled randomized clinical trial. Pain 2007, 127, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Akkoc, Y.; Uyar, M.; Oncu, J.; Ozcan, Z.; Durmaz, B. Complex regional pain syndrome in a patient with spinal cord injury: Management with pulsed radiofrequency lumbar sympatholysis. Spinal Cord 2008, 46, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Kane, T.P.; Rogers, P.; Hazelgrove, J.; Wimsey, S.; Harper, G.D. Pulsed radiofrequency applied to the suprascapular nerve in painful cuff tear arthropathy. J. Shoulder Elbow Surg. 2008, 17, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Zeldin, A.; Ioscovich, A. Pulsed radiofrequency for metastatic pain treatment. Pain Phys. 2008, 11, 921–922. [Google Scholar]
- Ramanavarapu, V.; Simopoulos, T.T. Pulsed radiofrequency of lumbar dorsal root ganglia for chronic post-amputation stump pain. Pain Phys. 2008, 11, 561–566. [Google Scholar]
- Sluijter, M.E.; van Kleef, M.; Barendse, G.A.; Weber, W. Thermal transport during radiofrequency current therapy of the intervertebral disc. Spine 1998, 23, 745. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, J.; de Louw, A.J.; Joosten, E.A.; Kessels, A.G.; Honig, W.; Dederen, P.J.; Veening, J.G.; Vles, J.S.; van Kleef, M. Pulsed and continuous radiofrequency current adjacent to the cervical dorsal root ganglion of the rat induces late cellular activity in the dorsal horn. Anesthesiology 2005, 102, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Cahana, A.; Vutskits, L.; Muller, D. Acute differential modulation of synaptic transmission and cell survival during exposure to pulsed and continuous radiofrequency energy. J. Pain 2003, 4, 197–202. [Google Scholar] [CrossRef]
- Protasoni, M.; Reguzzoni, M.; Sangiorgi, S.; Reverberi, C.; Borsani, E.; Rodella, L.F.; Dario, A.; Tomei, G.; Dell’Orbo, C. Pulsed radiofrequency effects on the lumbar ganglion of the rat dorsal root: A morphological light and transmission electron microscopy study at acute stage. Eur. Spine J. 2009, 18, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.R.; Gereau, R.W., 4th; Malcangio, M.; Strichartz, G.R. MAP kinase and pain. Brain Res. Rev. 2009, 60, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Crown, E.D. The role of mitogen activated protein kinase signaling in microglia and neurons in the initiation and maintenance of chronic pain. Exp. Neurol. 2012, 234, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, Y.; Satoh, Y.; Kodama, M.; Araki, Y.; Satomoto, M.; Sakamoto, E.; Pagès, G.; Pouysségur, J.; Endo, S.; Kazama, T. Mechanical allodynia but not thermal hyperalgesia is impaired in mice deficient for ERK2 in the central nervous system. Pain 2012, 153, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.L.; Lin, W.T.; Huang, R.Y.; Chen, T.C.; Huang, S.H.; Chang, C.H.; Tsai, S.Y.; Chiu, H.W.; Yeh, G.C.; Lin, C.W.; et al. Pulsed radiofrequency inhibited activation of spinal mitogen-activated protein kinases and ameliorated early neuropathic pain in rats. Eur. J. Pain 2014, 18, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.C.; Wu, Z.F.; Chen, J.C.; Wong, C.S.; Huang, C.J.; Wang, J.S.; Chien, C.C. Association between extracellular signal-regulated kinase expression and the anti-allodynic effect in rats with spared nerve injury by applying immediate pulsed radiofrequency. BMC Anesthesiol. 2015, 15, 92. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.R. Mitogen-activated protein kinases as potential targets for pain killers. Curr. Opin. Investig. Drugs 2004, 5, 71–75. [Google Scholar] [PubMed]
- Ma, W.; Quirion, R. The ERK/MAPK pathway, as a target for the treatment of neuropathic pain. Expert Opin. Ther. Targets 2005, 9, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, T.; Mizoguchi, H.; Sasaki, M.; Sakurada, C.; Tsuzuki, M.; Sakurada, S.; Sakurada, T. Inhibition of ERK phosphorylation by substance P N-terminal fragment decreases capsaicin-induced nociceptive response. Neuropharmacology 2011, 61, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Aksu, R.; Ugur, F.; Bicer, C.; Menku, A.; Guler, G.; Madenoglu, H.; Canpolat, D.G.; Boyaci, A. The efficiency of pulsed radiofrequency application on L5 and L6 dorsal roots in rabbits developing neuropathic pain. Reg. Anesth. Pain Med. 2010, 35, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Perret, D.M.; Kim, D.S.; Li, K.W.; Sinavsky, K.; Newcomb, R.L.; Miller, J.M.; Luo, Z.D. Application of pulsed radiofrequency currents to rat dorsal root ganglia modulates nerve injury-induced tactile allodynia. Anesth. Analg. 2011, 113, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Park, H.W.; Ahn, S.H.; Son, J.Y.; Kim, S.J.; Hwang, S.J.; Cho, Y.W.; Lee, D.G. Pulsed radiofrequency application reduced mechanical hypersensitivity and microglial expression in neuropathic pain model. Pain Med. 2012, 13, 1227–1234. [Google Scholar] [CrossRef]
- Van Boxem, K.; Huntoon, M.; van Zundert, J.; Patijn, J.; van Kleef, M.; Joosten, E.A. Pulsed radiofrequency: A review of the basic science as applied to the pathophysiology of radicular pain: A call for clinical translation. Reg. Anesth. Pain Med. 2014, 39, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, D.; Kostopanagiotou, G.; Lemonis, A.; Batistaki, C. Intradiscal combination of pulsed radiofrequency and gelified ethanol for the treatment of chronic discogenic low back pain. Pain Med. 2014, 15, 881–883. [Google Scholar] [CrossRef] [PubMed]
- Shanthanna, H.; Chan, P.; McChesney, J.; Thabane, L.; Paul, J. Pulsed radiofrequency treatment of the lumbar dorsal root ganglion in patients with chronic lumbar radicular pain: A randomized, placebo-controlled pilot study. J. Pain Res. 2014, 7, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, H.; Shao, H.; Xue, Q.; Yu, B. ERK MAP kinase activation in spinal cord regulates phosphorylation of Cdk5 at serine 159 and contributes to peripheral inflammation induced pain/hypersensitivity. PLoS ONE 2014, 9, 87788. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, A.; Flaman, A.S.; Prasad, S.S.; Gravel, C.; Williams, A.; Yauk, C.L.; Li, X. Acetaminophen modulates the transcriptional response to recombinant interferon-beta. PLoS ONE 2010, 5, 11031. [Google Scholar] [CrossRef] [PubMed]
- Gruber, H.E.; Hoelscher, G.L.; Ingram, J.A.; Hanley, E.N., Jr. Genome-wide analysis of pain-, nerve- and neurotrophin -related gene expression in the degenerating human annulus. Mol. Pain 2012, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Jin, E.H.; Zhang, E.; Ko, Y.; Sim, W.S.; Moon, D.E.; Yoon, K.J.; Hong, J.H.; Lee, W.H. Genome-wide expression profiling of complex regional pain syndrome. PLoS ONE 2013, 8, 79435. [Google Scholar] [CrossRef] [PubMed]
- Thibault, K.; Calvino, B.; Rivals, I.; Marchand, F.; Dubacq, S.; McMahon, S.B.; Pezet, S. Molecular mechanisms underlying the enhanced analgesic effect of oxycodone compared to morphine in chemotherapy-induced neuropathic pain. PLoS ONE 2014, 9, 91297. [Google Scholar] [CrossRef] [PubMed]
- Bello, M.; Leone, P.; Nebreda, P.; Kusak, M.; Decampos, J.; Vaquero, J.; Sarasa, J.; Pestana, A.; Rey, J. Molecular abnormalities of chromosome-19 in malignant gliomas—preferential involvement of the 19q13.2–q13.4 region. Int. J. Oncol. 1995, 6, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, Y.; Banno, Y.; Shimizu, Y.; Ando, S.; Hasegawa, H.; Adachi, K.; Iwamoto, T. Reduced adult hippocampal neurogenesis and working memory deficits in the Dgcr8-deficient mouse model of 22q11.2 deletion-associated schizophrenia can be rescued by IGF2. J. Neurosci. 2013, 33, 9408–9419. [Google Scholar] [CrossRef] [PubMed]
- Agis-Balboa, R.C.; Arcos-Diaz, D.; Wittnam, J.; Govindarajan, N.; Blom, K.; Burkhardt, S.; Haladyniak, U.; Agbemenyah, H.Y.; Zovoilis, A.; Salinas-Riester, G.; et al. A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J. 2011, 30, 4071–4083. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Gagneur, J.; Robinson, P.N. GOing Bayesian: Model-based gene set analysis of genome-scale data. Nucleic Acids Res. 2010, 38, 3523–3532. [Google Scholar] [CrossRef] [PubMed]
- Schmeisser, M.J.; Baumann, B.; Johannsen, S.; Vindedal, G.F.; Jensen, V.; Hvalby, O.C.; Sprengel, R.; Seither, J.; Maqbool, A.; Magnutzki, A.; et al. IkappaB kinase/nuclear factor kappaB-dependent insulin-like growth factor 2 (Igf2) expression regulates synapse formation and spine maturation via Igf2 receptor signaling. J. Neurosci. 2012, 32, 5688–5703. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Gao, F.; Tian, W.; Ruteshouser, E.C.; Wang, Y.; Lazar, A.; Stewart, J.; Strong, L.C.; Behringer, R.R.; Huff, V. Wt1 ablation and Igf2 upregulation in mice result in Wilms tumors with elevated ERK1/2 phosphorylation. J. Clin. Investig. 2011, 121, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Kuner, R. Central mechanisms of pathological pain. Nat. Med. 2010, 16, 1258–1266. [Google Scholar] [CrossRef] [PubMed]
- Rahn, E.J.; Guzman-Karlsson, M.C.; David Sweatt, J. Cellular, molecular, and epigenetic mechanisms in non-associative conditioning: Implications for pain and memory. Neurobiol. Learn. Mem. 2013, 105, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.Y.; Stern, S.A.; Garcia-Osta, A.; Saunier-Rebori, B.; Pollonini, G.; Bambah-Mukku, D.; Blitzer, R.D.; Alberini, C.M. A critical role for IGF-II in memory consolidation and enhancement. Nature 2011, 469, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.S.; Zhao, M.L.; Derico, L.; Choi, N.; Lee, S.C. Insulin-like growth factor 1 and 2 (IGF1, IGF2) expression in human microglia: Differential regulation by inflammatory mediators. J. Neuroinflamm. 2013, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.H.; Zhang, G.H.; Jia, D.; Wang, P.; Qian, N.S.; He, F.; Zeng, X.T.; He, Y.; Yang, Y.L.; Cao, D.Y.; et al. Spinal astrocytic activation contributes to mechanical allodynia in a mouse model of type 2 diabetes. Brain Res. 2011, 1368, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Abejon, D.; Garcia-del-Valle, S.; Fuentes, M.L.; Gomez-Arnau, J.I.; Reig, E.; van Zundert, J. Pulsed radiofrequency in lumbar radicular pain: Clinical effects in various etiological groups. Pain Pract. 2007, 7, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.J.; Korostynski, M.; Kaminska-Chowaniec, D.; Obara, I.; Mika, J.; Przewlocka, B.; Przewlocki, R. Comparison of gene expression profiles in neuropathic and inflammatory pain. J. Physiol. Pharmacol. 2006, 57, 401–414. [Google Scholar] [PubMed]
- Mika, J.; Osikowicz, M.; Rojewska, E.; Korostynski, M.; Wawrzczak-Bargiela, A.; Przewlocki, R.; Przewlocka, B. Differential activation of spinal microglial and astroglial cells in a mouse model of peripheral neuropathic pain. Eur. J. Pharmacol. 2009, 623, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Coyle, D.E. Partial peripheral nerve injury leads to activation of astroglia and microglia which parallels the development of allodynic behavior. Glia 1998, 23, 75–83. [Google Scholar] [CrossRef]
- Mika, J.; Rojewska, E.; Makuch, W.; Przewlocka, B. Minocycline reduces the injury-induced expression of prodynorphin and pronociceptin in the dorsal root ganglion in a rat model of neuropathic pain. Neuroscience 2010, 165, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.Y.; Gerner, P.; Woolf, C.J.; Ji, R.R. ERK is sequentially activated in neurons, microglia, and astrocytes by spinal nerve ligation and contributes to mechanical allodynia in this neuropathic pain model. Pain 2005, 114, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Mika, J.; Zychowska, M.; Popiolek-Barczyk, K.; Rojewska, E.; Przewlocka, B. Importance of glial activation in neuropathic pain. Eur. J. Pharmacol. 2013, 716, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Deumens, R.; Jaken, R.J.; Knaepen, L.; van der Meulen, I.; Joosten, E.A. Inverse relation between intensity of GFAP expression in the substantia gelatinosa and degree of chronic mechanical allodynia. Neurosci. Lett. 2009, 452, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Decosterd, I.; Woolf, C.J. Spared nerve injury: An animal model of persistent peripheral neuropathic pain. Pain 2000, 87, 149–158. [Google Scholar] [CrossRef]
- Tóth, Z.E.; Mezey, E. Simultaneous visualization of multiple antigens with tyramide signal amplification using antibodies from the same species. J. Histochem. Cytochem. 2007, 55, 545–554. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeh, C.-C.; Sun, H.-L.; Huang, C.-J.; Wong, C.-S.; Cherng, C.-H.; Huh, B.K.; Wang, J.-S.; Chien, C.-C. Long-Term Anti-Allodynic Effect of Immediate Pulsed Radiofrequency Modulation through Down-Regulation of Insulin-Like Growth Factor 2 in a Neuropathic Pain Model. Int. J. Mol. Sci. 2015, 16, 27156-27170. https://doi.org/10.3390/ijms161126013
Yeh C-C, Sun H-L, Huang C-J, Wong C-S, Cherng C-H, Huh BK, Wang J-S, Chien C-C. Long-Term Anti-Allodynic Effect of Immediate Pulsed Radiofrequency Modulation through Down-Regulation of Insulin-Like Growth Factor 2 in a Neuropathic Pain Model. International Journal of Molecular Sciences. 2015; 16(11):27156-27170. https://doi.org/10.3390/ijms161126013
Chicago/Turabian StyleYeh, Chun-Chang, Hsiao-Lun Sun, Chi-Jung Huang, Chih-Shung Wong, Chen-Hwan Cherng, Billy Keon Huh, Jinn-Shyan Wang, and Chih-Cheng Chien. 2015. "Long-Term Anti-Allodynic Effect of Immediate Pulsed Radiofrequency Modulation through Down-Regulation of Insulin-Like Growth Factor 2 in a Neuropathic Pain Model" International Journal of Molecular Sciences 16, no. 11: 27156-27170. https://doi.org/10.3390/ijms161126013






