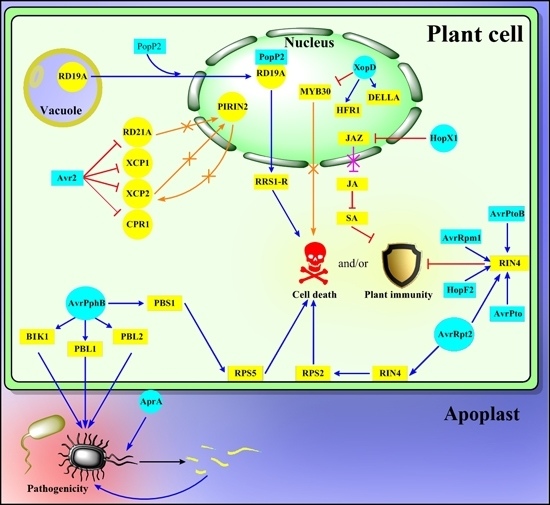
Regulatory Proteolysis in Arabidopsis-Pathogen Interactions
Abstract
:1. Introduction
| Gene | Full Name | AGI Code | Uniprot Accession | MEROPS Identifier | Reference |
|---|---|---|---|---|---|
| AtCDR1 | Constitutive Disease Resistance 1 | At5g33340 | Q6XBF8 | A01.069 | [14,15] |
| AtAED1 | Apoplastic EDS1-Dependent 1 | At5g10760 | Q9LEW3 | A01.A14 | [16] |
| AtRD21a | Responsive to Dehydration 21a | At1g47128 | P43297 | C01.064 | [17,18,19,20] |
| AtRD21b | Responsive to Dehydration 21b | At5g43060 | Q0WM94 | C01.A12 | [20] |
| AtXCP1 | Xylem Cysteine Proteinase 1 | At4g35350 | O65493 | C01.065 | [17] |
| AtXCP2 | Xylem Cysteine Proteinase 2 | At1g20850 | Q9LM66 | C01.120 | [17,20] |
| AtCPR1 | Probable Cysteine Proteinase | At3g19400 | Q9LT77 | C01.A12 | [17] |
| AtALEU | Aleurain | At5g60360 | Q8H166 | C01.163 | [17] |
| AtALEUL | Aleurain-Like | At3g45310 | Q8RWQ9 | C01.162 | |
| AtRD19a | Responsive to Dehydration 19a | At4g39090 | P43296 | C01.022 | [21] |
| AtCathB1 | Cathepsin B1, B2, B3 | At1g02300 | Q56XY7 | C01.A10 | [22] |
| AtCathB2 | At1g02305 | Q93VC9 | C01.144 | ||
| AtCathB3 | At4g01610 | Q9ZSI0 | C01.144 | ||
| AtMC1 | Metacaspase 1 | At1g02170 | Q7XJE6 | C14.047 | [23] |
| AtMC2 | Metacaspase 2 | At4g25110 | Q7XJE5 | C14.A04 | [23] |
| AtMC4 | Metacaspase 4 | At1g79340 | O64517 | C14.033 | [24] |
| AtαVPE | α, β, δ, or γ Vacuolar Processing Enzyme | At2g25940 | P49047 | C13.002 | [25,26,27,28] |
| AtβVPE | At1g62710 | Q39044 | C13.001 | ||
| AtδVPE | At3g20210 | Q9LJX8 | C13.A01 | ||
| AtγVPE | At4g32940 | Q39119 | C13.006 | ||
| AtCEP1 | KDEL Cysteine Endopeptidase 1 | At5g50260 | Q9FGR9 | C01.A03 | [29] |
| AtSBT3.3 | Subtilase 3.3 | At1g32960 | Q9MAP5 | S08.A35 | [30] |
| AtPBA1 | 26S Proteasome β1 Subunit | At4g31300 | Q8LD27 | T01.010 | [31] |
| Protease | Species | Uniprot Accession | MEROPS Identifier | Reference |
|---|---|---|---|---|
| AvrPphB | P. syringae pv. phaseolicola | Q52430 | C58.002 | [32,33,34] |
| AvrRpt2 | P. syringae pv. tomato | Q6LAD6 | C70.001 | [35,36,37,38,39,40,41] |
| XopD | X. campestris pv. vesicatoria | Q3BYJ5 | C48.023 | [42,43] |
| HopX1 | P. syringae pv. tabaci | Q83YM6 | N/A | [44] |
| protease IV | P. aeruginosa | Q02SZ7 | S01.281 | [45] |
| AprA | P. syringae pv. tomato | Q87ZU2 | M10.060 | [46] |
2. Functions of Proteolytic Enzymes in Various Arabidopsis thaliana Pathosystems
2.1. Host-Derived Aspartic Proteases
2.2. Host-Derived Cysteine Proteases
2.3. Host-Derived Serine Protease
2.4. Arabidopsis PBA1, the β1 Subunit of the 26S Proteasome
2.5. Cysteine Protease Effectors Secreted by Pathogens of Arabidopsis
2.6. Protease IV, a Bacterial Lysyl Class Serine Protease Effector
2.7. Alkaline Protease AprA, a Bacterial Zinc Metalloprotease
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. Merops: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014, 42, D503–D509. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoorn, R.A.L. Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223. [Google Scholar] [CrossRef] [PubMed]
- Tsiatsiani, L.; van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P.V. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.; Turk, D.; Turk, V. Protease signalling: The cutting edge. EMBO J. 2012, 31, 1630–1643. [Google Scholar] [CrossRef] [PubMed]
- Dunn, B.M. Determination of protease mechanism. In Proteolytic Enzymes: A Practical Approach, 2nd ed.; Beynon, R., Bond, J.S., Eds.; Oxford University Press: Oxford, UK, 2001; pp. 77–104. [Google Scholar]
- Duplan, V.; Rivas, S. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 2014, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Peeters, N.; Rivas, S. Ubiquitination during plant immune signaling. Plant Physiol. 2012, 160, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Furlan, G.; Klinkenberg, J.; Trujillo, M. Regulation of plant immune receptors by ubiquitination. Front. Plant Sci. 2012, 3, 238. [Google Scholar] [CrossRef] [PubMed]
- Dielen, A.-S.; Badaoui, S.; Candresse, T.; German-Retana, S. The ubiquitin/26S proteasome system in plant–pathogen interactions: A never-ending hide-and-seek game. Mol. Plant Pathol. 2010, 11, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Banfield, M.J. Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell. Microbiol. 2015, 17, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Hara-Nishimura, I. Two vacuole-mediated defense strategies in plants. Plant Signal. Behav. 2010, 5, 1568–1570. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Xia, Y.; Cameron, R.; Shadle, G.; Blount, J.; Lamb, C.; Dixon, R.A. Signals for local and systemic responses of plants to pathogen attack. J. Exp. Bot. 2004, 55, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Breitenbach, H.H.; Wenig, M.; Wittek, F.; Jorda, L.; Maldonado-Alconada, A.M.; Sarioglu, H.; Colby, T.; Knappe, C.; Bichlmeier, M.; Pabst, E.; et al. Contrasting roles of the apoplastic aspartyl protease apoplastic, enhanced disease susceptibility1-dependent1 and legume lectin-like protein1 in Arabidopsis systemic acquired resistance. Plant Physiol. 2014, 165, 791–809. [Google Scholar] [CrossRef]
- Van Esse, P.; van’t Klooster, J.W.; Bolton, M.D.; Yadeta, K.A.; van Baarlen, P.; Boeren, S.; Vervoort, J.; de Wit, P.J.G.M.; Thomma, B.P.H.J. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 2008, 20, 1948–1963. [Google Scholar] [CrossRef] [PubMed]
- Shindo, T.; Misas-Villamil, J.C.; Horger, A.C.; Song, J.; van der Hoorn, R.A.L. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS ONE 2012, 7, e29317. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fabregas, J.; Diaz-Moreno, I.; Gonzalez-Arzola, K.; Janocha, S.; Navarro, J.A.; Hervas, M.; Bernhardt, R.; Diaz-Quintana, A.; de la Rosa, M.A. New Arabidopsis thaliana cytochrome c partners: A look into the elusive role of cytochrome c in programmed cell death in plants. Mol. Cell. Proteom. 2013, 12, 3666–3676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tremousaygue, D.; Denance, N.; van Esse, H.P.; Horger, A.C.; Dabos, P.; Goffner, D.; Thomma, B.P.H.J.; van der Hoorn, R.A.L.; Tuominen, H. PIRIN2 stabilizes cysteine protease XCP2 and increases susceptibility to the vascular pathogen Ralstonia solanacearum in Arabidopsis. Plant J. 2014, 79, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Bernoux, M.; Timmers, T.; Jauneau, A.; Briere, C.; de Wit, P.J.; Marco, Y.; Deslandes, L. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 2008, 20, 2252–2264. [Google Scholar] [CrossRef] [PubMed]
- McLellan, H.; Gilroy, E.M.; Yun, B.-W.; Birch, P.R.J.; Loake, G.J. Functional redundancy in the Arabidopsis Cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytol. 2009, 183, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 2011, 66, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Kuroyanagi, M.; Yamada, K.; Hatsugai, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 2005, 280, 32914–32920. [Google Scholar] [CrossRef] [PubMed]
- Rojo, E.; Martin, R.; Carter, C.; Zouhar, J.; Pan, S.; Plotnikova, J.; Jin, H.; Paneque, M.; Sanchez-Serrano, J.J.; Baker, B.; et al. VPEγ exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 2004, 14, 1897–1906. [Google Scholar] [CrossRef] [PubMed]
- Misas-Villamil, J.C.; Toenges, G.; Kolodziejek, I.; Sadaghiani, A.M.; Kaschani, F.; Colby, T.; Bogyo, M.; van der Hoorn, R.A.L. Activity profiling of vacuolar processing enzymes reveals a role for VPE during oomycete infection. Plant J. 2013, 73, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Qiang, X.; Zechmann, B.; Reitz, M.U.; Kogel, K.-H.; Schafer, P. The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress–triggered caspase-dependent cell death. Plant Cell 2012, 24, 794–809. [Google Scholar] [CrossRef] [PubMed]
- Höwing, T.; Huesmann, C.; Hoefle, C.; Nagel, M.-K.; Isono, E.; Hückelhoven, R.; Gietl, C. Endoplasmic reticulum KDEL-tailed cysteine endopeptidase 1 of Arabidopsis (AtCEP1) is involved in pathogen defense. Front. Plant Sci. 2015, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Ramırez, V.; Lopez, A.; Mauch-Mani, B.; Gil, M.J.; Vera, P. An extracellular subtilase switch for immune priming in Arabidopsis. PLoS Pathog. 2013, 9, e1003445. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Iwasaki, S.; Tamura, K.; Kondo, M.; Fuji, K.; Ogasawara, K.; Nishimura, M.; Hara-Nishimura, I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Gene Dev. 2009, 23, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Golstein, C.; Ade, J.; Stoutemyer, M.; Dixon, J.E.; Innes, R.W. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 2003, 301, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- Ade, J.; DeYoung, B.J.; Golstein, C.; Innes, R.W. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 2007, 104, 2531–2536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, B.N.; Bent, A.F.; Dahlbeck, D.; Innes, R.W.; Staskawicz, B.J. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 1993, 5, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Chisholm, S.T.; Dahlbeck, D.; Staskawicz, B.J. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 2003, 49, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Staskawicz, B.J. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 2003, 112, 369–377. [Google Scholar] [CrossRef]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Kim, H.-S.; Desveaux, D.; Singer, A.U.; Patel, P.; Sondek, J.; Dangl, J.L. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA 2005, 102, 6496–6501. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.J.; da Cunha, L.; Mackey, D. Separable fragments and membrane tethering of Arabidopsis RIN4 regulate its suppression of PAMP-triggered immunity. Plant Cell 2011, 23, 3798–3811. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Wu, S.; Sun, W.; Coaker, G.; Kunkel, B.; He, P.; Shan, L. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 2013, 162, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Canonne, J.; Marino, D.; Jauneau, A.; Pouzet, C.; Briere, C.; Roby, D.; Rivas, S. The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell 2011, 23, 3498–3511. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.M.; Li, M.-Y.; Yang, P.-Y.; Chang, S.H.; Ho, Y.-P.; Lin, H.; Deng, W.L.; Yang, J.Y. Arabidopsis HFR1 is a potential nuclear substrate regulated by the Xanthomonas type III effector XopDXcc8004. PLoS ONE 2015, 10, e0117067. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Boter, M.; Fernandez-Barbero, G.; Chini, A.; Rathjen, J.P.; Solano, R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014, 12, e1001792. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Li, J.-F.; Niu, Y.; Zhang, X.-C.; Woody, O.Z.; Xiong, Y.; Djonovic, S.; Millet, Y.; Bush, J.; McConkey, B.J.; et al. Pathogen-secreted proteases activate a novel plant immune pathway. Nature 2015, 521, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Pel, M.J.C.; van Dijken, A.J.H.; Bardoel, B.W.; Seidl, M.F.; van der Ent, S.; van Strijp, J.A.G.; Pieterse, C.M.J. Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol. Plant Microbe Interact. 2014, 27, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Simoes, I.; Faro, R.; Bur, D.; Faro, C. Characterization of recombinant CDR1, an Arabidopsis aspartic proteinase involved in disease resistance. J. Biol. Chem. 2007, 282, 31358–31365. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoorn, R.A.L.; Leeuwenburgh, M.A.; Bogyo, M.; Joosten, M.H.A.J.; Peck, S.C. Activity profiling of papain-like cysteine proteases in plants. Plant Physiol. 2004, 135, 1170–1178. [Google Scholar] [CrossRef] [PubMed]
- Richau, K.H.; van der Hoorn, R.A.L. Studies on plant-pathogen interactions using activity-based proteomics. Curr. Proteom. 2010, 7, 328–336. [Google Scholar] [CrossRef]
- Van der Hoorn, R.A.L.; Kaiser, M. Probes for activity-based profiling of plant proteases. Physiol. Plant. 2012, 145, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Rooney, H.C.; van’t Klooster, J.W.; van der Hoorn, R.A.L.; Joosten, M.H.A.J.; Jones, J.D.G.; de Wit, P.J.G.M. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 2005, 308, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, M.; Yamaguchi-Shinozaki, K.; Tsuji, H.; Shinozaki, K. Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 1993, 129, 175–182. [Google Scholar] [CrossRef]
- Krishnan, R.V.; Masuda, A.; Centonze, V.E.; Herman, B. Quantitative imaging of protein-protein interactions by multiphoton fluorescence lifetime imaging microscopy using a streak camera. J. Biomed. Opt. 2003, 8, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.; Stoka, V. Protease signalling in cell death: Caspases versus cysteine cathepsins. FEBS Lett. 2007, 581, 2761–2767. [Google Scholar] [CrossRef] [PubMed]
- Wrzaczek, M.; Vainonen, J.P.; Stael, S.; Tsiatsiani, L.; Help-Rinta-Rahko, H.; Gauthier, A.; Kaufholdt, D.; Bollhöner, B.; Lamminmäki, A.; Staes, A.; et al. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 2015, 34, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Yamada, K.; Goto-Yamada, S.; Hara-Nishimura, I. Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 2015, 6, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hierl, G.; Vothknecht, U.; Gietl, C. Programmed cell death in Ricinus and Arabidopsis: The function of KDEL cysteine peptidases in development. Physiol. Plant. 2012, 145, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Stintzi, A.; Graff, L. Subtilases—Versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant. 2012, 145, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Genov, N.; Shopova, M.; Boteva, R.; Jori, G.; Richelli, F. Chemical, photochemical and spectroscopic characterization of an alkaline proteinase from Bacillus subtilis variant DY. Biochem. J. 1982, 207, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Chichkova, N.V.; Shaw, J.; Galiullina, R.A.; Drury, G.E.; Tuzhikov, A.I.; Kim, S.H.; Kalkum, M.; Hong, T.B.; Gorshkova, E.N.; Torrance, L.; et al. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J. 2010, 29, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Vartapetian, A.B.; Tuzhikov, A.I.; Chichkova, N.V.; Taliansky, M.; Wolpert, T.J. A plant alternative to animal caspases: Subtilisin-like proteases. Cell Death Differ. 2011, 18, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Fu, H.; Walker, J.; Papa, C.M.; Smalle, J.; Ju, Y.M.; Vierstra, R.D. Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 2004, 279, 6401–6413. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Smalle, J.A. Structure, function and regulation of plant proteasomes. Biochimie 2008, 90, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Brentnall, M.; Rodriguez-Menocal, L.; de Guevara, R.L.; Cepero, E.; Boise, L.H. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 2013, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Simonich, M.T.; Innes, R.W. A disease resistance gene in Arabidopsis with specificity for the avrPph3 gene of Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 1995, 8, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, L.; Rivas, S. Catch me if you can: Bacterial effectors and plant targets. Trends Plant Sci. 2012, 17, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Hotson, A.; Chosed, R.; Shu, H.; Orth, K.; Mudgett, M.B. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 2003, 50, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Daniel, X.; Lacomme, C.; Morel, J.-B.; Roby, D. A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J. 1999, 20, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Vailleau, F.; Daniel, X.; Tronchet, M.; Montillet, J.L.; Triantaphylides, C.; Roby, D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. USA 2002, 99, 10179–10184. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Vailleau, F.; Leger, A.; Joubes, J.; Miersch, O.; Huard, C.; Blee, E.; Mongrand, S.; Domergue, F.; Roby, D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 2008, 20, 752–767. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Rong, W.; Luo, H.; Chen, Y.; He, C. The Xanthomonas campestris effector protein XopDXcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 2014, 204, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Gene Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef] [PubMed]
- Dill, A.; Jung, H.-S.; Sun, T.-P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 2001, 98, 14162–14167. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cheng, T.; Wang, P.; Tian, L.; Wang, G.; Luo, Y.; Wang, J.; Yang, L.; Shi, J. Genome-wide bioinformatics analysis of DELLA-family proteins from plants. Plant Omics 2013, 6, 201–207. [Google Scholar]
- Sun, T. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010, 154, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Bari, R.; Achard, P.; Lison, P.; Nemri, A.; Harberd, N.P.; Jones, J.D.G. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 2008, 18, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Da Cunha, L.; Sreerekha, M.-V.; Mackey, D. Defense suppression by virulence effectors of bacterial phytopathogens. Curr. Opin. Plant Biol. 2007, 10, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Bocsanczy, A.M.; Schneider, D.J.; DeClerck, G.A.; Cartinhour, S.; Beer, S.V. HopX1 in Erwinia amylovora functions as an avirulence protein in apple and is regulated by HrpL. J. Bacteriol. 2012, 194, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Bahrami, A.K.; Pringle, E.G.; Hernandez-Guzman, G.; Bender, C.L.; Pierce, N.E.; Ausubel, F.M. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA 2005, 102, 1791–1796. [Google Scholar] [PubMed]
- Laurie-Berry, N.; Joardar, V.; Street, I.H.; Kunkel, B.N. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol. Plant Microbe Interact. 2006, 19, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, S.; Shinoda, S. Microbial metalloproteases and pathogenesis. Microbes Infect. 2000, 2, 91–98. [Google Scholar] [CrossRef]
- Létoffé, S.; Delepelaire, P.; Wandersman, C. Characterization of a protein inhibitor of extracellular proteases produced by Erwinia chrysanthemi. Mol. Microbiol. 1989, 3, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Duong, F.; Lazdunski, A.; Cami, B.; Murgier, M. Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: Relationships to other secretory pathways. Gene 1992, 12, 47–54. [Google Scholar] [CrossRef]
- Guzzo, J.; Duong, F.; Wandersman, C.; Murgier, M.; Lazdunski, A. The secretion genes of Pseudomonas aeruginosa alkaline protease are functionally related to those of Erwinia chrysanthemi proteases and Escherichia coli alpha-haemolysin. Mol. Microbiol. 1991, 5, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Buell, C.R.; Joardar, V.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Gwinn, M.L.; Dodson, R.J.; Deboy, R.T.; Durkin, A.S.; Kolonay, J.F.; et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2003, 100, 10181–10186. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.; Pham, X.; Erwin, A.; Mizoguchi, S.; Warrener, P.; Hickey, M.; Brinkman, F.; Hufnagle, W.; Kowalik, D.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [PubMed]
- Bardoel, B.W.; van der Ent, S.; Pel, M.J.C.; Tommassen, J.; Pieterse, C.M.J.; van Kessel, K.P.M.; van Strijp, J.A.G. Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog. 2011, 7, e1002206. [Google Scholar] [CrossRef] [PubMed]
- Belenghi, B.; Acconcia, F.; Trovato, M.; Perazzolli, M.; Bocedi, A.; Polticelli, F.; Ascenzi, P.; Delledonne, M. AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur. J. Biochem. 2003, 270, 2593–2604. [Google Scholar] [CrossRef] [PubMed]
- Lampl, N.; Alkan, N.; Davydov, O.; Fluhr, R. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 2013, 74, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Dickman, M.B.; Fluhr, R. Centrality of host cell death in plant-microbe interactions. Annu. Rev. Phytopathol. 2013, 51, 543–570. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.M.; dos Reis, S.P.; de Souza, C.R. Phytocystatins and their potential to control plant diseases caused by fungi. Protein Pept. Lett. 2015, 22, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.-T. Kunitz trypsin inhibitor: An antagonist of cell death triggered by phytopathogens and fumonisin b1 in Arabidopsis. Mol. Plant 2008, 1, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Shindo, T.; van der Hoorn, R.A.L. Papain-like cysteine proteases: Key players at molecular battlefields employed by both plants and their invaders. Mol. Plant Pathol. 2008, 9, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Szatmari, A.; Zvara, A.; Moricz, A.M.; Besenyei, E.; Szabo, E.; Ott, P.G.; Puskas, L.G.; Bozso, Z. Pattern triggered immunity (PTI) in tobacco: Isolation of activated genes suggests role of the phenylpropanoid pathway in inhibition of bacterial pathogens. PLoS ONE 2014, 9, e102869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.J.; Kroon, J.T.M.; Simon, W.J.; Slabas, A.R.; Chivasa, S. A novel function for Arabidopsis CYCLASE1 in programmed cell death revealed by iTRAQ analysis of extracellular matrix proteins. Mol. Cell. Proteom. 2015, 14, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pogány, M.; Dankó, T.; Kámán-Tóth, E.; Schwarczinger, I.; Bozsó, Z. Regulatory Proteolysis in Arabidopsis-Pathogen Interactions. Int. J. Mol. Sci. 2015, 16, 23177-23194. https://doi.org/10.3390/ijms161023177
Pogány M, Dankó T, Kámán-Tóth E, Schwarczinger I, Bozsó Z. Regulatory Proteolysis in Arabidopsis-Pathogen Interactions. International Journal of Molecular Sciences. 2015; 16(10):23177-23194. https://doi.org/10.3390/ijms161023177
Chicago/Turabian StylePogány, Miklós, Tamás Dankó, Evelin Kámán-Tóth, Ildikó Schwarczinger, and Zoltán Bozsó. 2015. "Regulatory Proteolysis in Arabidopsis-Pathogen Interactions" International Journal of Molecular Sciences 16, no. 10: 23177-23194. https://doi.org/10.3390/ijms161023177