Identification and Analysis of Differentially-Expressed microRNAs in Japanese Encephalitis Virus-Infected PK-15 Cells with Deep Sequencing
Abstract
:1. Introduction
2. Results and Discussion
2.1. Overview of the Deep Sequencing Results
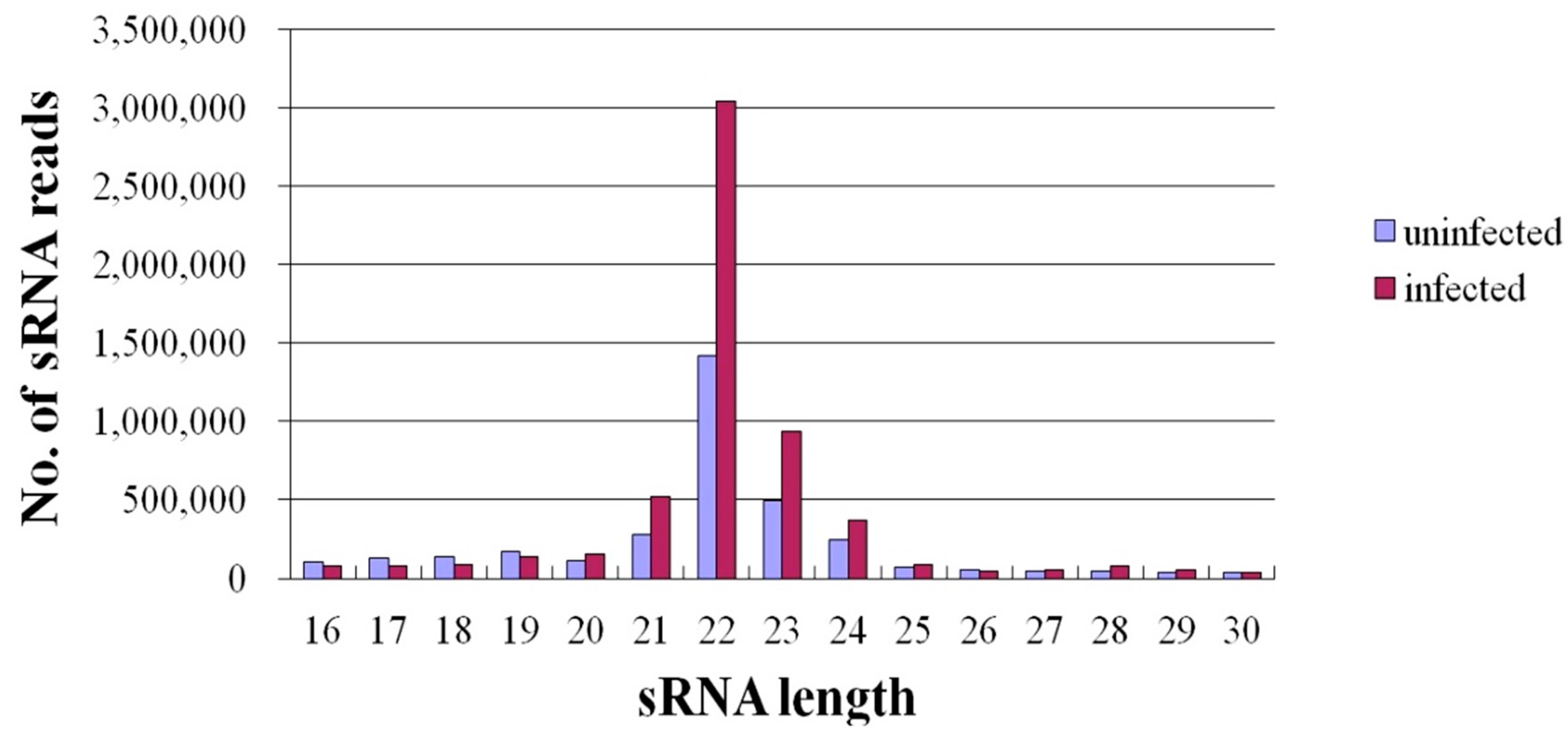
2.2. Expression of miRNAs in JEV-Infected and -Uninfected PK-15 Cells
| Ranking | JEV-Infected Group | JEV-Uninfected Group | ||
|---|---|---|---|---|
| miRNA Reads | miRNA Reads | |||
| 1 | ssc-miR-21 | 17,39,040 | ssc-miR-21 | 877,629 |
| 2 | ssc-let-7f | 309,868 | ssc-let-7f | 151,697 |
| 3 | ssc-miR-30a-5p | 69,597 | ssc-miR-19b | 33,441 |
| 4 | ssc-miR-100 | 60,186 | ssc-miR-24-3p | 23,501 |
| 5 | ssc-miR-29a | 53,334 | ssc-miR-152 | 22,650 |
| 6 | ssc-miR-152 | 49,317 | ssc-miR-18a | 21,872 |
| 7 | ssc-miR-10a-5p | 39,632 | ssc-let-7a | 20,908 |
| 8 | ssc-miR-19b | 37,389 | ssc-miR-100 | 16,274 |
| 9 | ssc-miR-26a | 35,650 | ssc-miR-19a | 14,533 |
| 10 | ssc-miR-182 | 29,255 | ssc-miR-30a-5p | 14,489 |
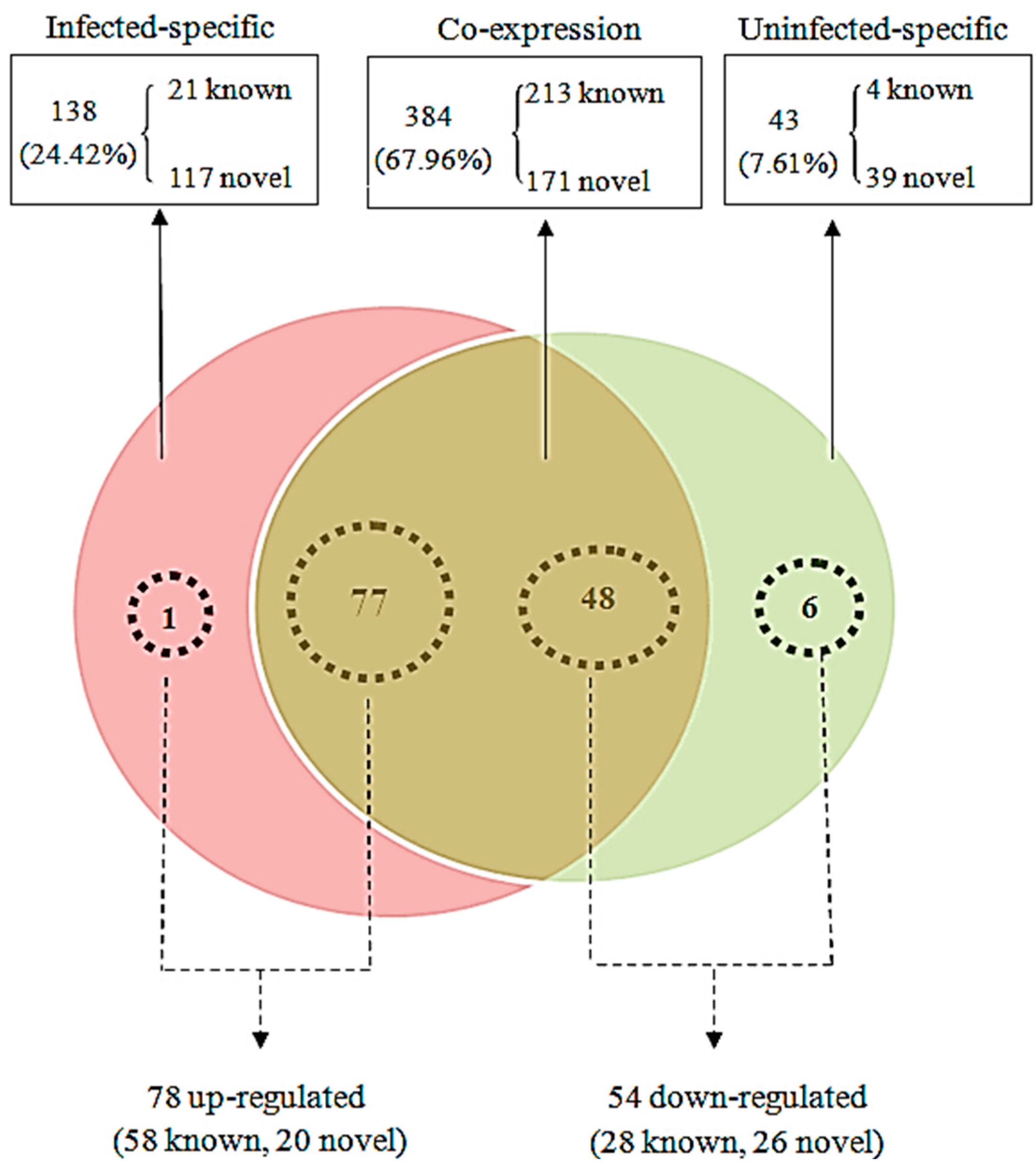
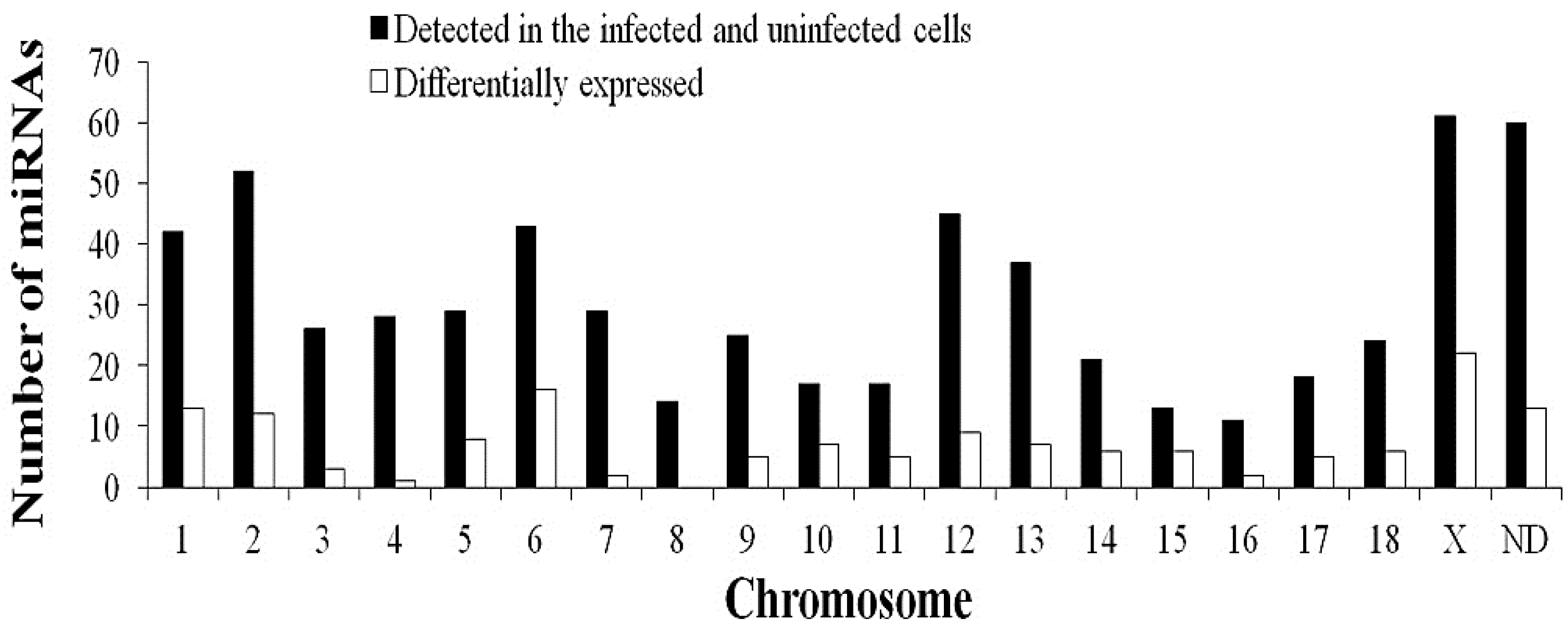
2.3. Differentially-Expressed miRNAs in JEV-Infected PK-15 Cells
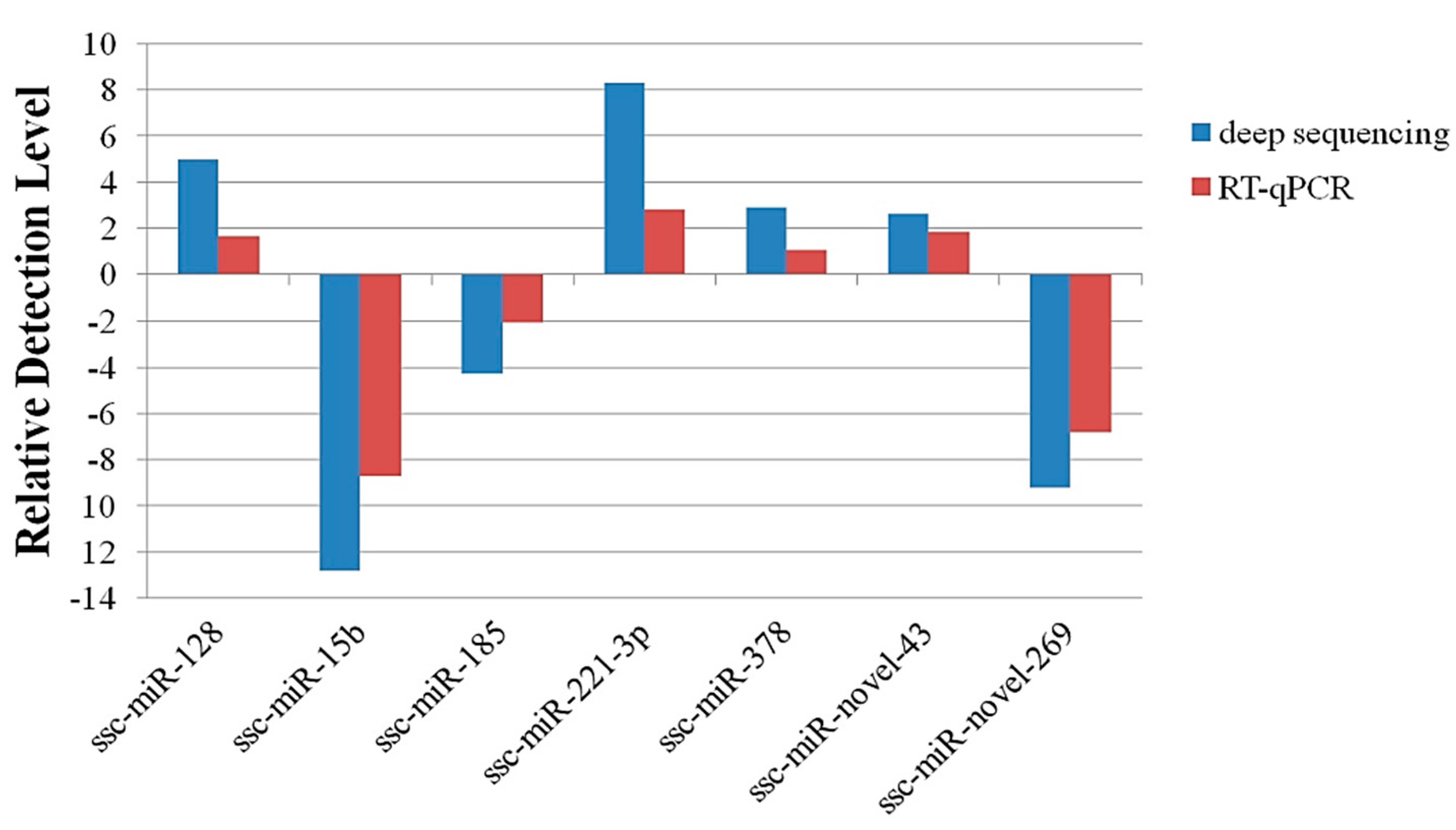
2.4. Validation of Deep Sequencing Results by RT-qPCR
| Primer | Sequence (5'-3') |
|---|---|
| ssc-miR-128-forward | TCACAGTGAACCGGTCTCTTT |
| ssc-miR-15b-forward | TAGCAGCACATCATGGTTTACA |
| ssc-miR-185-forward | TGGAGAGAAAGGCAGTTCCTGA |
| ssc-miR-221-3p-forward | AGCTACATTGTCTGCTGGGTTT |
| ssc-miR-378-forward | ACTGGACTTGGAGTCAGAAGGC |
| ssc-miR-novel-43-forward | TTCAAGTAACCCAGGATAGGCT |
| ssc-miR-novel-269-forward | TACCCATTGCATATCGGAGTTG |
| miR-reverse | GTCGGTGTCGTGGAGTCG |
| U6-forward | TCGCTTTGGCAGCACCTAT |
| U6-reverse | AATATGGAACGCTTCGCAAA |
| Poly(T) adapter | GTCGGTGTCGTGGAGTCGTTTGCAATTGCACTGGATTTTTTTTTTTTTTTTTTV |
2.5. GO Analysis of Predicted Targets of Differentially-Expressed miRNAs

3. Experimental Section
3.1. Cells and Virus
3.2. RNA Isolation and Illumina Sequencing
3.3. Analysis of Sequencing Data
3.4. Identification of Differentially-Expressed miRNAs
3.5. Reverse Transcription-Quantitative Real-Time PCR (RT-qPCR)
3.6. miRNA Target Prediction
3.7. Gene Ontology Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Erlanger, T.E.; Weiss, S.; Keiser, J.; Utzinger, J.; Wiedenmayer, K. Past, present, and future of Japanese encephalitis. Emerg. Infect. Dis. 2009, 15, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hollidge, B.S.; González-Scarano, F.; Soldan, S.S. Arboviral encephalitides: Transmission, emergence, and pathogenesis. J. Neuroimmune Pharmacol. 2010, 5, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Nasci, R.; Liang, G. The neglected arboviral infections in mainland China. PLoS Negl. Trop. Dis. 2010, 4, e624. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, J.; Boqvist, S.; Ståhl, K.; Thu, H.T.V.; Magnusson, U. Reproductive performance in sows in relation to Japanese Encephalitis Virus seropositivity in an endemic area. Trop. Anim. Health Prod. 2012, 44, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Meister, G. miRNAs get an early start on translational silencing. Cell 2007, 131, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Martello, G.; Piccolo, S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Schickel, R.; Boyerinas, B.; Park, S.; Peter, M. MicroRNAs: Key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008, 27, 5959–5974. [Google Scholar] [CrossRef] [PubMed]
- Scaria, V.; Hariharan, M.; Maiti, S.; Pillai, B.; Brahmachari, S.K. Host-virus interaction: A new role for microRNAs. Retrovirology 2006, 3, 68. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, R.; Jeang, K.-T. The roles of microRNAs in mammalian virus infection. Biochim. Biophys. Acta 2008, 1779, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, T.; Wakita, T.; Yang, W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology 2010, 398, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Anselmo, A.; Flori, L.; Jaffrezic, F.; Rutigliano, T.; Cecere, M.; Cortes-Perez, N.; Lefevre, F.; Rogel-Gaillard, C.; Giuffra, E. Co-expression of host and viral microRNAs in porcine dendritic cells infected by the pseudorabies virus. PLoS One 2011, 6, e17374. [Google Scholar] [CrossRef] [PubMed]
- Pareek, S.; Roy, S.; Kumari, B.; Jain, P.; Banerjee, A.; Vrati, S. miR-155 induction in microglial cells suppresses Japanese encephalitis virus replication and negatively modulates innate immune responses. J. Neuroinflamm. 2014, 11, 97. [Google Scholar] [CrossRef]
- Thounaojam, M.C.; Kundu, K.; Kaushik, D.K.; Swaroop, S.; Mahadevan, A.; Shankar, S.K.; Basu, A. MicroRNA 155 regulates Japanese encephalitis virus-induced inflammatory response by targeting Src homology 2-containing inositol phosphatase 1. J. Virol. 2014, 88, 4798–4810. [Google Scholar] [CrossRef] [PubMed]
- Thounaojam, M.C.; Kaushik, D.K.; Kundu, K.; Basu, A. MicroRNA-29b modulates Japanese encephalitis virus-induced microglia activation by targeting tumor necrosis factor α-induced protein 3. J. Neurochem. 2014, 129, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2009, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Buermans, H.P.; Ariyurek, Y.; van Ommen, G.; den Dunnen, J.T.; ACʼt Hoen, P. New methods for next generation sequencing based microRNA expression profiling. BMC Genomics 2010, 11, 716. [Google Scholar] [CrossRef]
- Dutta, K.; Rangarajan, P.N.; Vrati, S.; Basu, A. Japanese encephalitis: Pathogenesis, prophylactics and therapeutics. Curr. Sci. 2010, 98, 326–334. [Google Scholar]
- Liu, K.; Liao, X.; Zhou, B.; Yao, H.; Fan, S.; Chen, P.; Miao, D. Porcine α interferon inhibit Japanese encephalitis virus replication by different ISGs in vitro. Res. Vet. Sci. 2013, 95, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; He, M.; Liu, X.; Li, X.; Fan, B.; Zhao, S. Japanese encephalitis virus infects porcine kidney epithelial PK15 cells via clathrin-and cholesterol-dependent endocytosis. Virol. J. 2013, 10, 258. [Google Scholar] [CrossRef]
- Fan, J.-M.; Luo, J.; Chen, L.; Teng, M.; Bu, D.; Wang, F.-Y.; Wang, L.; Wang, C.-Q.; Zhang, G.-P. Genetic analysis of strains of Japanese Encephalitis Virus isolated from swine in central China. Virus Genes 2010, 40, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.S.; Tyagi, S.; Nancarrow, D.J.; Boyle, G.M.; Cook, A.L.; Whiteman, D.C.; Parsons, P.G.; Schmidt, C.; Sturm, R.A.; Hayward, N.K. Characterization of the melanoma miRNAome by deep sequencing. PLoS One 2010, 5, e9685. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Y.; Wang, T.; Guan, J.; Luo, Z.; Chen, H.; Wang, X.; Chen, L.; Ma, J.; Mu, Z. Repertoire of porcine microRNAs in adult ovary and testis by deep sequencing. Int. J. Biol. Sci. 2011, 7, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.A.; Yoo, D.; Liu, H.-C. Characterization of the microRNAome in Porcine Reproductive and Respiratory Syndrome Virus Infected Macrophages. PLoS One 2013, 8, e82054. [Google Scholar] [CrossRef] [PubMed]
- Buggele, W.A.; Johnson, K.E.; Horvath, C.M. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J. Biol. Chem. 2012. [Google Scholar] [CrossRef]
- Li, S.-C.; Chan, W.-C.; Ho, M.-R.; Tsai, K.-W.; Hu, L.-Y.; Lai, C.-H.; Hsu, C.-N.; Hwang, P.-P.; Lin, W.-C. Discovery and characterization of medaka miRNA genes by next generation sequencing platform. BMC Genomics 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Cao, L.; Xu, Z.; Fang, L.; Zhong, Y.; Chen, Q.; Luo, R.; Chen, H.; Li, K.; Xiao, S. MiR-125b reduces porcine reproductive and respiratory syndrome virus replication by negatively regulating the NF-κB pathway. PLoS One 2013, 8, e55838. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Hou, J.; Lin, L.; Wang, C.; Liu, X.; Li, D.; Ma, F.; Wang, Z.; Cao, X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 2010, 185, 6226–6233. [Google Scholar] [CrossRef] [PubMed]
- Varnholt, H.; Drebber, U.; Schulze, F.; Wedemeyer, I.; Schirmacher, P.; Dienes, H.P.; Odenthal, M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 2008, 47, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-H.; Wang, B.; Kota, J.; Yu, J.; Costinean, S.; Kutay, H.; Yu, L.; Bai, S.; la Perle, K.; Chivukula, R.R. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Investig. 2012, 122, 2871–2883. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Nerurkar, V.R. Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology 2014, 452, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Wu, D.; Li, P.; Xu, B.; Lu, Q.; Zhang, W. microRNA-18a, a member of the oncogenic miR-17–92 cluster, targets Dicer and suppresses cell proliferation in bladder cancer T24 cells. Mol. Med. Rep. 2012, 5, 167–172. [Google Scholar] [PubMed]
- Triboulet, R.; Mari, B.; Lin, Y.-L.; Chable-Bessia, C.; Bennasser, Y.; Lebrigand, K.; Cardinaud, B.; Maurin, T.; Barbry, P.; Baillat, V. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 2007, 315, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, M.; Jing, Q.; Georgel, P.; New, L.; Chen, J.; Mols, J.; Kang, Y.J.; Jiang, Z.; Du, X.; Cook, R. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 2007, 27, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Cobb, B.S.; Hertweck, A.; Smith, J.; O'connor, E.; Graf, D.; Cook, T.; Smale, S.T.; Sakaguchi, S.; Livesey, F.J.; Fisher, A.G. A role for Dicer in immune regulation. J. Exp. Med. 2006, 203, 2519–2527. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D.; Boldin, M.P.; O'Connell, R.M.; Rao, D.S.; Taganov, K.D. MicroRNAs: New regulators of immune cell development and function. Nat. Immunol. 2008, 9, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Trenkmann, M.; Brock, M.; Gay, R.E.; Michel, B.A.; Gay, S.; Huber, L.C. Tumor necrosis factor α-induced microRNA-18a activates rheumatoid arthritis synovial fibroblasts through a feedback loop in NF-κB signaling. Arthritis Rheumatol. 2013, 65, 916–927. [Google Scholar] [CrossRef]
- Brock, M.; Trenkmann, M.; Gay, R.E.; Gay, S.; Speich, R.; Huber, L.C. MicroRNA-18a enhances the interleukin-6-mediated production of the acute-phase proteins fibrinogen and haptoglobin in human hepatocytes. J. Biol. Chem. 2011, 286, 40142–40150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, V.; Mathur, A.; Krishnani, N.; Dhole, T.N. An insufficient anti-inflammatory cytokine response in mouse brain is associated with increased tissue pathology and viral load during Japanese encephalitis virus infection. Arch. Virol. 2008, 153, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dutta, K.; Kumawat, K.L.; Ghoshal, A.; Adhya, D.; Basu, A. Abrogated inflammatory response promotes neurogenesis in a murine model of Japanese encephalitis. PLoS One 2011, 6, e17225. [Google Scholar] [CrossRef] [PubMed]
- Pauley, K.M.; Chan, E.K. MicroRNAs and their emerging roles in immunology. Ann. N. Y. Acad. Sci. 2008, 1143, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.-Y. The role of microRNAs in regulatory T cells and in the immune response. Immun. Netw. 2011, 11, 11–41. [Google Scholar] [CrossRef]
- Ye, J.; Zhu, B.; Fu, Z.F.; Chen, H.; Cao, S. Immune evasion strategies of flaviviruses. Vaccine 2013, 31, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Meng, Q.; Li, Y.; Xiu, Y.; Du, J.; Gu, W.; Wu, T.; Li, W.; Ding, Z.; Wang, W. Identification and comparative analysis of the Eriocheir sinensis microRNA transcriptome response to Spiroplasma eriocheiris infection using a deep sequencing approach. Fish Shellfish Immunol. 2012, 32, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.-Y.; Liu, Q.; Chen, J.-X.; Lan, K.; Ge, B.-X. MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. J. Immunol. 2010, 185, 7435–7442. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Q.; Song, Y.; Lai, L.; Wang, J.; Yu, H.; Cao, X.; Wang, Q. MicroRNA-98 negatively regulates IL-10 production and endotoxin tolerance in macrophages after LPS stimulation. FEBS Lett. 2011, 585, 1963–1968. [Google Scholar] [CrossRef] [PubMed]
- Tsukerman, P.; Enk, J.; Mandelboim, O. Metastamir-mediated immune evasion: miR-10b downregulates the stress-induced molecule MICB, hence avoid recognition by NKG2D receptor. Oncoimmunology 2013, 2, e22245. [Google Scholar] [CrossRef]
- Jin, R.; Zhu, W.; Cao, S.; Chen, R.; Jin, H.; Liu, Y.; Wang, S.; Wang, W.; Xiao, G. Japanese encephalitis virus activates autophagy as a viral immune evasion strategy. PLoS One 2013, 8, e52909. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.; Han, W.; Sui, X.; Xie, J.; Pan, H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015, 356, 332–338. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Y.; Zhu, L.; Zhou, Y.; Liu, X.; Liu, X.; Li, X.; Lang, Q.; Qiao, X.; Xu, Z. Identification and Analysis of Differentially-Expressed microRNAs in Japanese Encephalitis Virus-Infected PK-15 Cells with Deep Sequencing. Int. J. Mol. Sci. 2015, 16, 2204-2219. https://doi.org/10.3390/ijms16012204
Cai Y, Zhu L, Zhou Y, Liu X, Liu X, Li X, Lang Q, Qiao X, Xu Z. Identification and Analysis of Differentially-Expressed microRNAs in Japanese Encephalitis Virus-Infected PK-15 Cells with Deep Sequencing. International Journal of Molecular Sciences. 2015; 16(1):2204-2219. https://doi.org/10.3390/ijms16012204
Chicago/Turabian StyleCai, Yuhan, Ling Zhu, Yuanchen Zhou, Xiao Liu, Xiaowan Liu, Xinqiong Li, Qiaoli Lang, Xiaogai Qiao, and Zhiwen Xu. 2015. "Identification and Analysis of Differentially-Expressed microRNAs in Japanese Encephalitis Virus-Infected PK-15 Cells with Deep Sequencing" International Journal of Molecular Sciences 16, no. 1: 2204-2219. https://doi.org/10.3390/ijms16012204






