Surveillance and Cleavage of Eukaryotic tRNAs
Abstract
:1. Introduction
2. Eukaryotic Transfer RNA (tRNA) Turnover and Quality Control
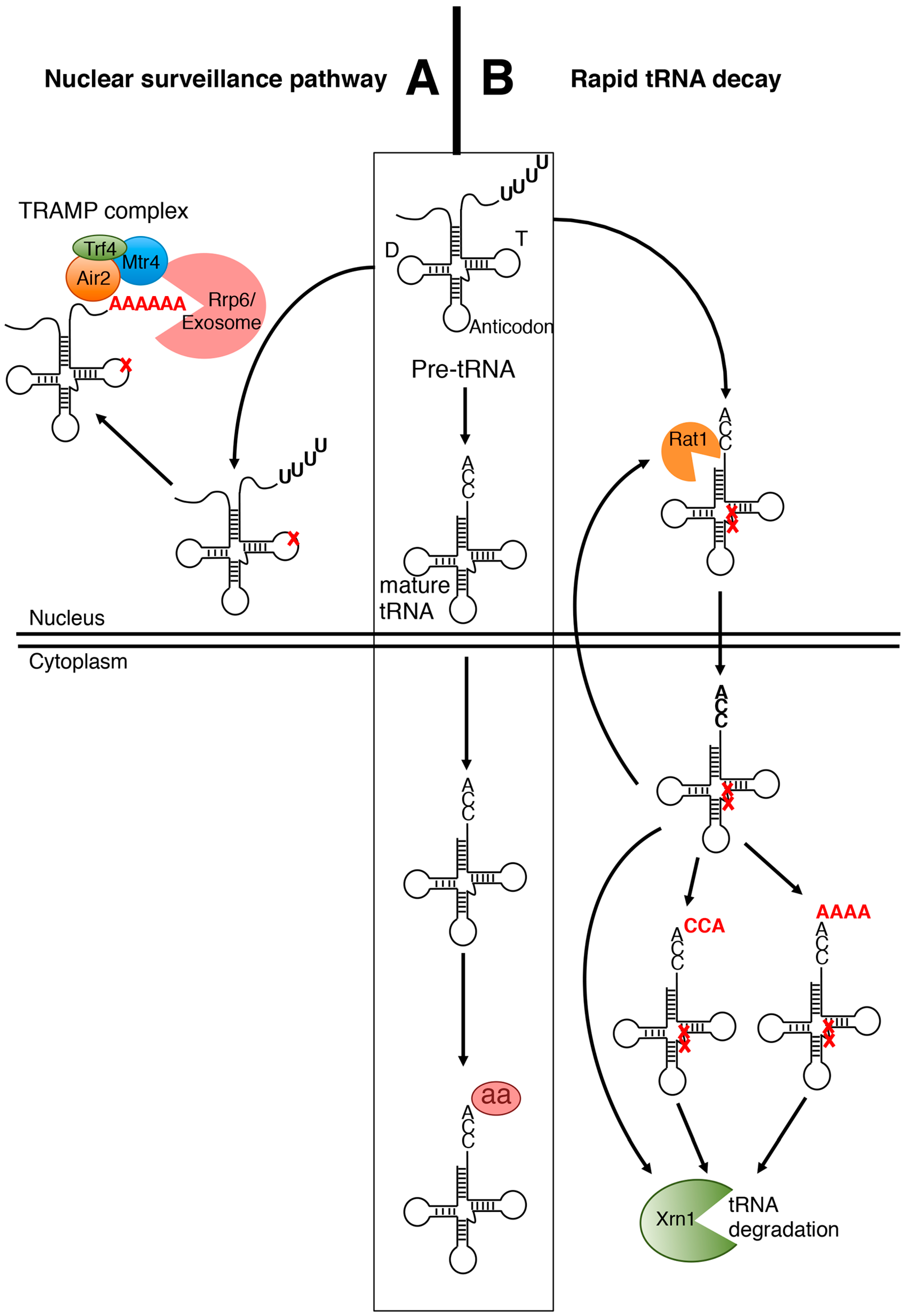
2.1. The Nuclear Surveillance Pathway
2.2. The Rapid tRNA Decay (RTD) Pathway
3. tRNA-Derived Fragments (tRFs): A New Class of Small Non-Coding RNAs
| tRFs | Yamasaki, 2009; Anderson, 2014 | Raina, 2014 | Lee, 2009 | General Nomenclature | |
|---|---|---|---|---|---|
 | 5'-tRF | 5'-tRF | tRF5 | tRF-5D | tRF-3D |
 | 3' CCA tRF | 3' CCA tRF | tRF3 | tRF-5T | tRF-3T |
 | 3' U tRF | tRF1 | pre-tRF-3U | ||
 | 5'-halves (5'-tiRNAs) | 5' tRNA halves | tRF-5A | tRF3A | |
 | 3'-halves (3'-tiRNAs) | 3' tRNA halves | tRF-5A | tRF3A | |
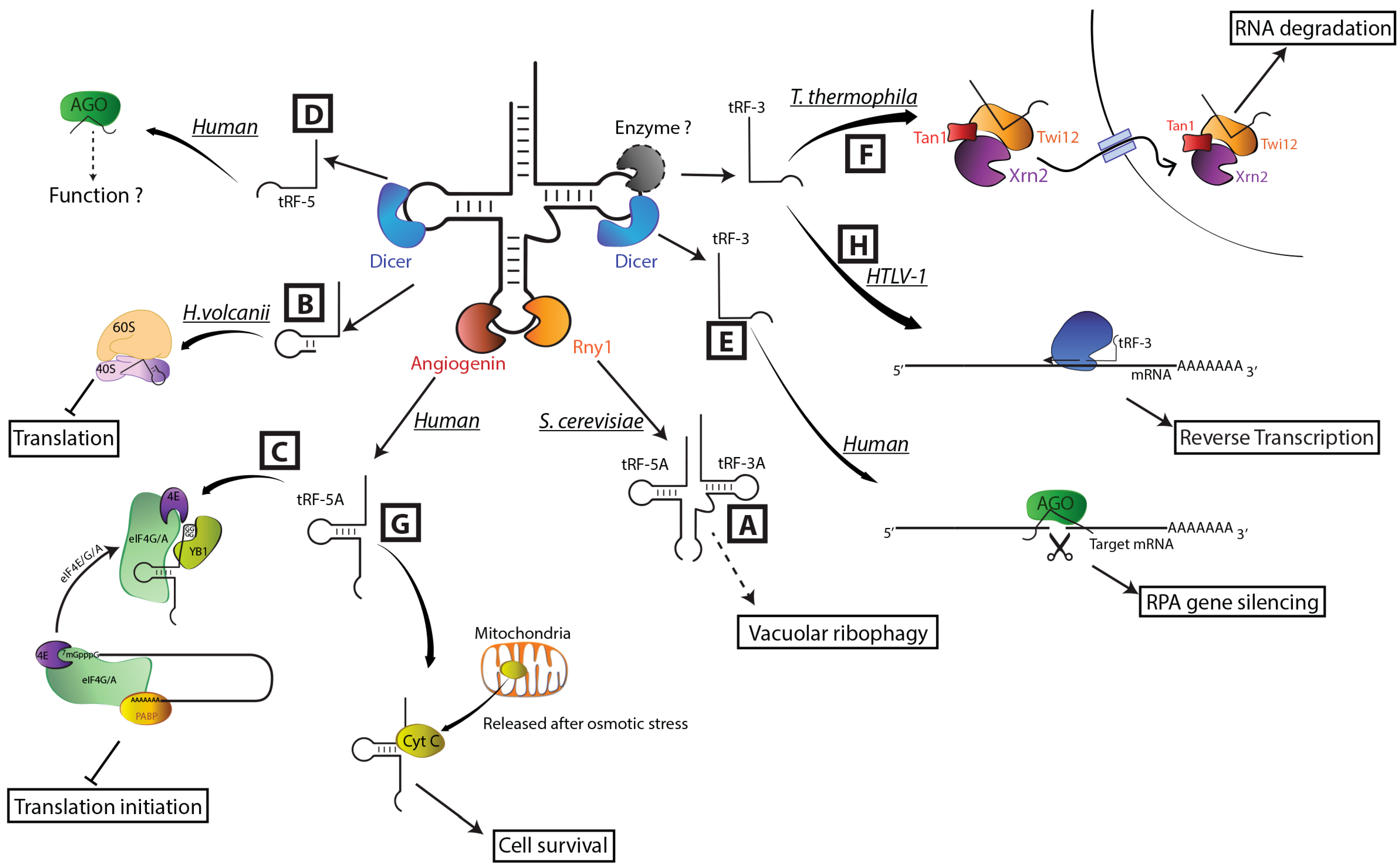
4. Biogenesis of tRF: Which Endonuclease for Which Job?
| Organisms | Endonucleases | tRNA Specificities | Cleavage Sites | References |
|---|---|---|---|---|
| E. coli | Colicin E5 | Queuine-containing tRNA; tRNAArg | Anticodon loop | [65] |
| Colicin D | Anticodon region | [66] | ||
| PrrC | tRNALys | Anticodon loop | [84] | |
| K. lactis | Zymocin | tRNALys; tRNAGln | Anticodon loop | [86] |
| S. cerevisiae | Rny1 | No | Anticodon loop | [87] |
| T. thermophila | Rnt2A–C | No | Anticodon loop | [90] |
| Mammals | ELAC2 | pre-tRNASer | 3' end | [52] |
| Dicer | tRNAGln | D loop | [73] | |
| tRNAGlu | T loop | [57] | ||
| pre-tRNAIle | 3' end | [91] | ||
| Angiogenin | No | Anticodon loop | [69] |
5. The Emerging Roles of tRFs
6. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Holley, R.; Apgar, J.; Everett, G.A.; Madison, J.T.; Marquisee, M.; Merril, S.H.; Penswick, J.R.; Zamir, A. Structure of a ribonucleic acid. Science 1965, 147, 1462–1465. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Quigley, G.J.; Suddath, F.L.; McPherson, A.; Sneden, D.; Kim, J.J.; Weinzierl, J.; Rich, A. Three-dimensional structure of yeast phenylalanine transfer RNA: Folding of the polynucleotide chain. Science 1973, 179, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Ladner, J.E.; Jack, A.; Robertus, J.D.; Brown, R.S.; Rhodes, D.; Clark, B.F.C.; Klug, A. Structure of yeast phenylalanine transfer RNA at 2.5 angström resolution. Proc. Natl. Acad. Sci. USA 1975, 72, 4414–4418. [Google Scholar] [CrossRef] [PubMed]
- Wende, S.; Platzer, E.G.; Juhling, F.; Putz, J.; Florentz, C.; Stadler, P.F.; Mörl, M. Biological evidence for the world’s smallest tRNAs. Biochimie 2013, 100, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Reznik, B.; Lykke-Andersen, J. Regulated and quality-control mRNA turnover pathways in eukaryotes. Biochem. Soc. Trans. 2010, 38, 1506–1510. [Google Scholar] [CrossRef] [PubMed]
- Parker, R. RNA degradation in Saccharomyces cerevisae. Genetics 2012, 191, 671–702. [Google Scholar] [CrossRef] [PubMed]
- Schoenberg, D.R.; Maquat, L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012, 13, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Grayhack, E.J.; Chernyakov, I.; Whipple, J.M. Roles of tRNA modifications in tRNA turnover. In DNA and RNA Modifiction Enzymes: Strucutre, Mechanism, Function and Evolution; Grosjean, H., Ed.; Landes Bioscience: Austin, TX, USA, 2009; pp. 564–576. [Google Scholar]
- Dare, K.; Ibba, M. Roles of tRNA in cell wall biosynthesis. Wiley Interdiscip. Rev. RNA 2012, 3, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Polacek, N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Raina, M.; Ibba, M. tRNAs as regulators of biological processes. Front. Genet. 2014, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Phan, L.; Cuesta, R.; Carlson, B.A.; Pak, M.; Asano, K.; Bjork, G.R.; Tamame, M.; Hinnebusch, A.G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998, 12, 3650–3662. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Phan, L.; Hinnebusch, A.G. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 5173–5178. [Google Scholar] [CrossRef] [PubMed]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Kadaba, S.; Krueger, A.; Trice, T.; Krecic, A.M.; Hinnebusch, A.G.; Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004, 18, 1227–1240. [Google Scholar] [CrossRef]
- Anderson, J.T. RNA turnover: Unexpected consequences of being tailed. Curr. Biol. 2005, 15, R635–R638. [Google Scholar] [CrossRef] [PubMed]
- Kadaba, S.; Wang, X.; Anderson, J.T. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5s rRNA. RNA 2006, 12, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jia, H.; Jankowsky, E.; Anderson, J.T. Degradation of hypomodified tRNAiMet in vivo involves RNA-dependent ATPase activity of the DEexH helicase Mtr4p. RNA 2008, 14, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Maraia, R.J.; Lamichhane, T.N. 3' Processing of eukaryotic precursor tRNAs. Wiley Interdiscip. Rev. RNA 2011, 2, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Vanacova, S.; Wolf, J.; Martin, G.; Blank, D.; Dettwiller, S.; Friedlein, A.; Langen, H.; Keith, G.; Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005, 3, e189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copela, L.A.; FeRNAndez, C.F.; Sherrer, R.L.; Wolin, S.L. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 2008, 14, 1214–1227. [Google Scholar] [CrossRef] [PubMed]
- Mörl, M.; Marchfelder, A. The final cut. The importance of tRNA 3'-processing. EMBO Rep. 2001, 2, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.J.; Wolin, S.L. The yeast La protein is required for the 3' endonucleolytic cleavage that matures tRNA precursors. Cell 1997, 89, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Ozanick, S.G.; Wang, X.; Costanzo, M.; Brost, R.L.; Boone, C.; Anderson, J.T. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2009, 37, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Kotelawala, L.; Grayhack, E.J.; Phizicky, E.M. Identification of yeast tRNA Um44 2'-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNASer species. RNA 2008, 14, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Dewe, J.M.; Whipple, J.M.; Chernyakov, I.; Jaramillo, L.N.; Phizicky, E.M. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 2012, 18, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.R.; Uhlenbeck, O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA 1988, 85, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Nobles, K.N.; Yarian, C.S.; Liu, G.; Guenther, R.H.; Agris, P.F. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2002, 30, 4751–4760. [Google Scholar] [CrossRef] [PubMed]
- Whipple, J.M.; Lane, E.A.; Chernyakov, I.; D’Silva, S.; Phizicky, E.M. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011, 25, 1173–1184. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Whipple, J.M.; Phizicky, E.M.; Sharp, P.A. tRNAs marked with CCACCA are targeted for degradation. Science 2011, 334, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Chernyakov, I.; Whipple, J.M.; Kotelawala, L.; Grayhack, E.J.; Phizicky, E.M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'–3' exonucleases Rat1 and Xrn1. Genes Dev. 2008, 22, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Miyagawa, R.; Tomikawa, C.; Mizuno, R.; Takahashi, A.; Hori, H.; Ijiri, K. Degradation of initiator tRNAMet by Xrn1/2 via its accumulation in the nucleus of heat-treated HeLa cells. Nucleic Acids Res. 2013, 41, 4671–4685. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, H.H.; Hopper, A.K. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2005, 102, 11290–11295. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, B.; Stevens, A.; Tollervey, D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997, 16, 7184–7195. [Google Scholar] [CrossRef] [PubMed]
- Petfalski, E.; Dandekar, T.; Henry, Y.; Tollervey, D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 1998, 18, 1181–1189. [Google Scholar] [PubMed]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef] [PubMed]
- Turowski, T.W.; Karkusiewicz, I.; Kowal, J.; Boguta, M. Maf1-mediated repression of RNA polymerase III transcription inhibits tRNA degradation via RTD pathway. RNA 2012, 18, 1823–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boguta, M.; Graczyk, D. RNA polymerase III under control: Repression and de-repression. Trends Biochem. Sci. 2011, 36, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Karkusiewicz, I.; Turowski, T.W.; Graczyk, D.; Towpik, J.; Dhungel, N.; Hopper, A.K.; Boguta, M. Maf1 protein, repressor of RNA polymerase III, indirectly affects tRNA processing. J. Biol. Chem. 2011, 286, 39478–39488. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Steitz, T.A. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 2004, 430, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Steitz, T.A. A story with a good ending: tRNA 3'-end maturation by CCA-adding enzymes. Curr. Opin. Struct. Biol. 2006, 16, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Lizano, E.; Scheibe, M.; Rammelt, C.; Betat, H.; Mörl, M. A comparative analysis of CCA-adding enzymes from human and E. coli: Differences in CCA addition and tRNA 3'-end repair. Biochimie 2008, 90, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Betat, H.; Rammelt, C.; Mörl, M. tRNA nucleotidyltransferases: Ancient catalysts with an unusual mechanism of polymerization. Cell. Mol. Life Sci. 2010, 67, 1447–1463. [Google Scholar] [CrossRef] [PubMed]
- Siwaszek, A.; Ukleja, M.; Dziembowski, A. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 2014. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Nakamura, M.; Takahashi, Y.; Sandelin, A.; Katayama, S.; Fukuda, S.; Daub, C.O.; Kai, C.; Kawai, J.; Yasuda, J.; et al. Hidden layers of human small RNAs. BMC Genomics 2008, 9, 157. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008, 135, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell 2003, 5, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Galizi, R.; Spano, F.; Giubilei, M.A.; Capuccini, B.; Magini, A.; Urbanelli, L.; Ogawa, T.; Dubey, J.P.; Spaccapelo, R.; Emiliani, C.; et al. Evidence of tRNA cleavage in apicomplexan parasites: Half-tRNAs as new potential regulatory molecules of Toxoplasma gondii and Plasmodium berghei. Mol. Biochem. Parasitol. 2013, 188, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Frugier, M.; Tosar, J.P.; Correa-Dominguez, A.; Ronalte-Alves, L.; Parodi-Talice, A.; Rovira, C.; Robello, C.; Goldenberg, S.; Cayota, A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol. Biochem. Parasitol. 2010, 171, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Zywicki, M.; Kunzi, A.; Polacek, N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 2012, 260909. [Google Scholar] [CrossRef] [PubMed]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed]
- Jochl, C.; Rederstorff, M.; Hertel, J.; Stadler, P.F.; Hofacker, I.L.; Schrettl, M.; Haas, H.; Huttenhofer, A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008, 36, 2677–2689. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luo, J.; Zhou, H.; Liao, J.Y.; Ma, L.M.; Chen, Y.Q.; Qu, L.H. Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008, 36, 6048–6055. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lee, I.; Ren, J.; Ajay, S.S.; Lee, Y.S.; Bao, X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 2013, 21, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Hayashizaki, Y. Exploration of small RNAs. PLoS Genet. 2008, 4, e22. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shi, J.; Zhang, Y.; Zhang, H.; Liao, S.; Li, W.; Lei, L.; Han, C.; Ning, L.; Cao, Y.; et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012, 22, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Tomita, K.; Ueda, T.; Watanabe, K.; Uozumi, T.; Masaki, H. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 1999, 283, 2097–2100. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Ogawa, T.; Uozumi, T.; Watanabe, K.; Masaki, H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl. Acad. Sci. USA 2000, 97, 8278–8283. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005, 280, 42744–42749. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Collins, K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006, 20, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Ivanov, P.; Hu, G.f.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Haiser, H.J.; Karginov, F.V.; Hannon, G.J.; Elliot, M.A. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008, 36, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.C.; Lin, S.I.; Kuo, H.F.; Chiou, T.J. Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots. Plant Signal. Behav. 2010, 5, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Yamakawa, A.; Boffelli, D.; Mote, P.; Martin, D.I. 5' tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics 2013, 14, 298. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.S.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, X.; Wang, H.; Lu, Y.Z.; de Ruiter, M.; Prins, M.; He, Y.K. A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa). BMC Genomics 2011, 12, 289. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals argonaute association with RNA fragments of diverse origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.Y.; Ma, L.M.; Guo, Y.H.; Zhang, Y.C.; Zhou, H.; Shao, P.; Chen, Y.Q.; Qu, L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3' trailers. PLoS One 2010, 5, e10563. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Krokowski, D.; Guan, B.J.; Ivanov, P.; Parisien, M.; Hu, G.F.; Anderson, P.; Pan, T.; Hatzoglou, M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 2012, 287, 42708–42725. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M. Circulating small noncoding RNAs as biomarkers of aging. Ageing Res. Rev. 2014, 17, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Nowacka, M.; Strozycki, P.M.; Jackowiak, P.; Hojka-Osinska, A.; Szymanski, M.; Figlerowicz, M. Identification of stable, high copy number, medium-sized RNA degradation intermediates that accumulate in plants under non-stress conditions. Plant Mol. Biol. 2013, 83, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Ivanov, P. tRNA fragments in human health and disease. FEBS Lett. 2014, 588, 4297–4304. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Hanada, T.; Weitzer, S.; Mair, B.; Bernreuther, C.; Wainger, B.J.; Ichida, J.; Hanada, R.; Orthofer, M.; Cronin, S.J.; Komnenovic, V.; et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 2013, 495, 474–480. [Google Scholar] [CrossRef] [PubMed]
- David, M.; Borasio, G.D.; Kaufmann, G. Bacteriophage T4-induced anticodon-loop nuclease detected in a host strain restrictive to RNA ligase mutants. Proc. Natl. Acad. Sci. USA 1982, 79, 7097–7101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanga-Kanfi, S.; Amitsur, M.; Azem, A.; Kaufmann, G. PrrC-anticodon nuclease: Functional organization of a prototypical bacterial restriction RNase. Nucleic Acids Res. 2006, 34, 3209–3219. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Esberg, A.; Huang, B.; Bystrom, A.S. Kluyveromyces lactis γ-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res. 2008, 36, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Luhtala, N.; Parker, R. Structure-function analysis of Rny1 in tRNA cleavage and growth inhibition. PLoS One 2012, 7, e41111. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Kawamata, T.; Horie, T.; Tsugawa, H.; Nakayama, Y.; Ohsumi, Y.; Fukusaki, E. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 2014. [Google Scholar] [CrossRef]
- Andersen, K.L.; Collins, K. Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate Tetrahymena. Mol. Biol. Cell 2011, 23, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hu, G.F. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J. Cell. Physiol. 2012, 227, 2822–2826. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. Stressing out over tRNA cleavage. Cell 2009, 138, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.M.; Ivanov, P.; Hickman, T.; Dawra, N.; Tisdale, S.; Kedersha, N.; Hu, G.F.; Anderson, P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010, 285, 10959–10968. [Google Scholar] [CrossRef] [PubMed]
- Czech, A.; Wende, S.; Mörl, M.; Pan, T.; Ignatova, Z. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet. 2013, 9, e1003767. [Google Scholar] [CrossRef] [PubMed]
- Reifur, L.; Garcia-Silva, M.R.; Poubel, S.B.; Alves, L.R.; Arauco, P.; Buiar, D.K.; Goldenberg, S.; Cayota, A.; Dallagiovanna, B. Distinct subcellular localization of tRNA-derived fragments in the infective metacyclic forms of Trypanosoma cruzi. Mem. Inst. Oswaldo Cruz 2012, 107, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011, 43, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Bounova, G.; Purdom, E.; Speed, T.P.; Collins, K. A Tetrahymena Piwi bound to mature tRNA 3' fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol. Cell 2012, 48, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Jobava, R.; Parisien, M.; Putnam, A.; Krokowski, D.; Gao, X.H.; Guan, B.J.; Yuan, Y.; Jankowsky, E.; Feng, Z.; et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell. Biol. 2014, 34, 2450–2463. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, K.; Guffanti, A.; Corradin, A.; Sharma, V.K.; De Bellis, G.; Corti, G.; Grassi, A.; Zanovello, P.; Bronte, V.; Ciminale, V.; et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: A role for a tRNA fragment as a primer for reverse transcriptase. J. Virol. 2014, 88, 3612–3622. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; O’Day, E.; Emara, M.M.; Wagner, G.; Lieberman, J.; Anderson, P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. USA 2014, 111, 18201–18206. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, X.; Liu, J.; Kiba, T.; Woo, J.; Ojo, T.; Hafner, M.; Tuschl, T.; Chua, N.H.; Wang, X.J. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J. 2011, 67, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Buhler, M.; Spies, N.; Bartel, D.P.; Moazed, D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 2008, 15, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Loss-Morais, G.; Waterhouse, P.M.; Margis, R. Description of plant tRNA-derived RNA fragments (tRFs) associated with Argonaute and identification of their putative targets. Biol. Direct 2013, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Durdevic, Z.; Mobin, M.B.; Hanna, K.; Lyko, F.; Schaefer, M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 2013, 4, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Lee, S.R.; Hogstad, B.; Malone, C.D.; Tonkin, L.A.; Sachidanandam, R.; Hannon, G.J.; Collins, K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009, 23, 2016–2032. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Sachidanandam, R.; Collins, K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010, 24, 2742–2747. [Google Scholar] [CrossRef] [PubMed]
- Hopper, A.K. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 2013, 194, 43–67. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Megel, C.; Morelle, G.; Lalande, S.; Duchêne, A.-M.; Small, I.; Maréchal-Drouard, L. Surveillance and Cleavage of Eukaryotic tRNAs. Int. J. Mol. Sci. 2015, 16, 1873-1893. https://doi.org/10.3390/ijms16011873
Megel C, Morelle G, Lalande S, Duchêne A-M, Small I, Maréchal-Drouard L. Surveillance and Cleavage of Eukaryotic tRNAs. International Journal of Molecular Sciences. 2015; 16(1):1873-1893. https://doi.org/10.3390/ijms16011873
Chicago/Turabian StyleMegel, Cyrille, Geoffrey Morelle, Stéphanie Lalande, Anne-Marie Duchêne, Ian Small, and Laurence Maréchal-Drouard. 2015. "Surveillance and Cleavage of Eukaryotic tRNAs" International Journal of Molecular Sciences 16, no. 1: 1873-1893. https://doi.org/10.3390/ijms16011873