Ectopic Expression of CsCTR1, a Cucumber CTR-Like Gene, Attenuates Constitutive Ethylene Signaling in an Arabidopsis ctr1-1 Mutant and Expression Pattern Analysis of CsCTR1 in Cucumber (Cucumis sativus)
Abstract
:1. Introduction
2. Results
2.1. Cloning of Open Reading Frame (ORF) of CsCTR1 and Sequence Analysis
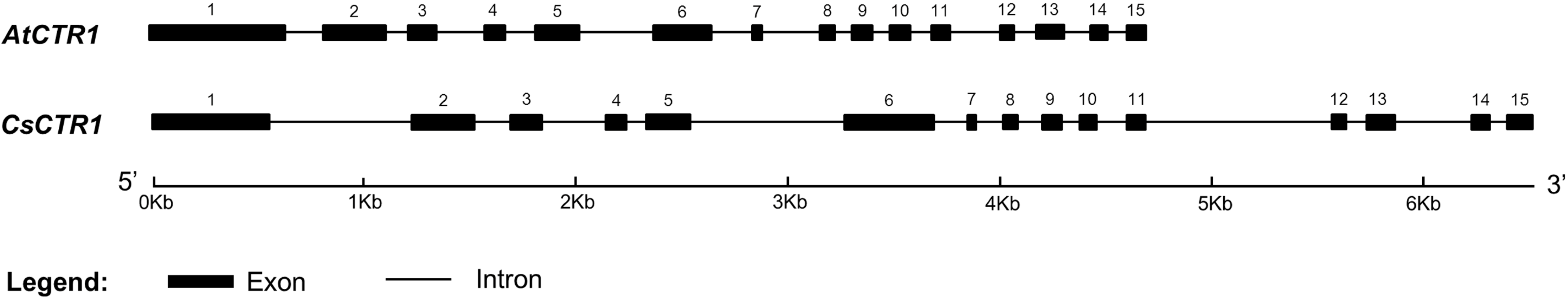
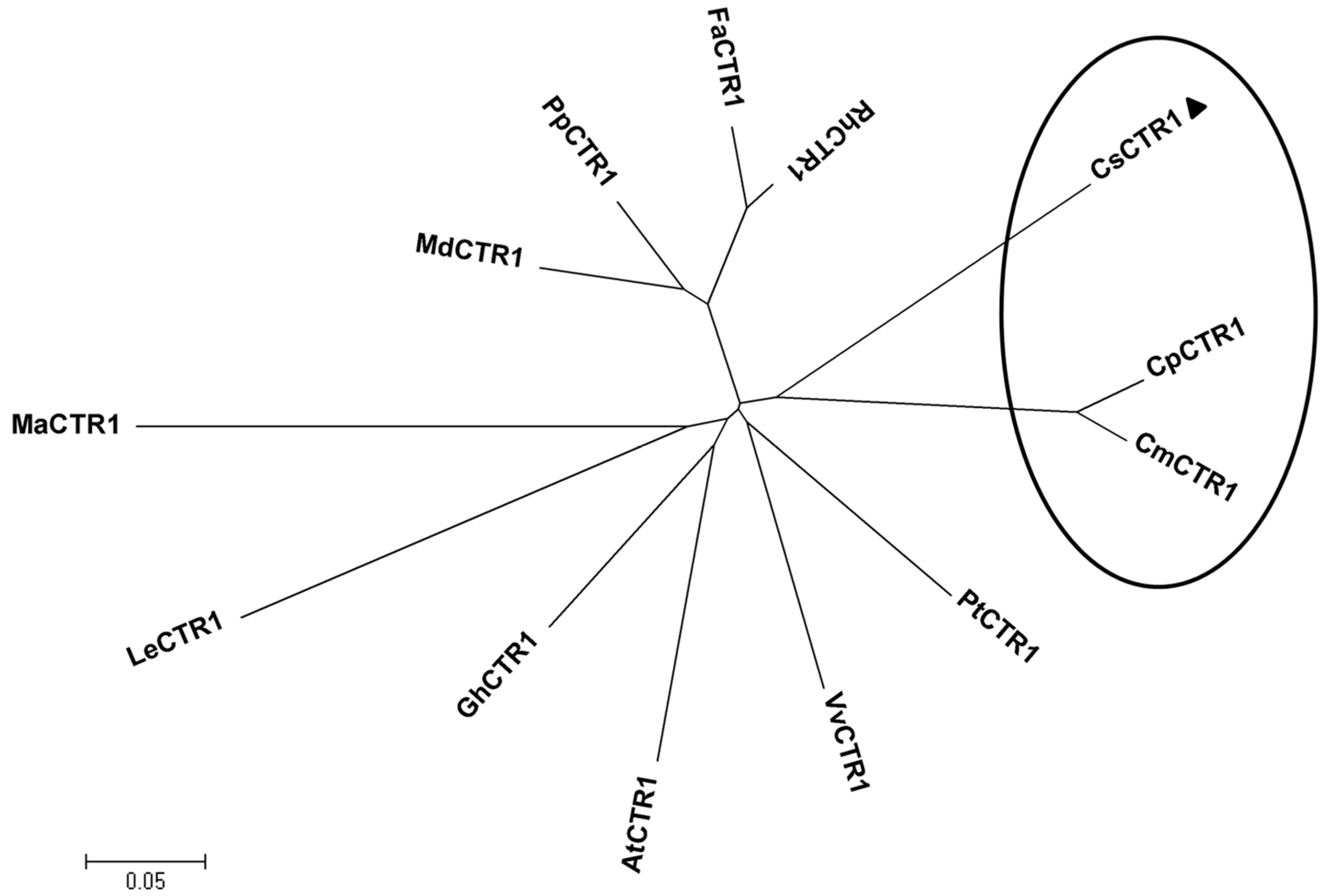
2.2. Multiple Sequence Alignment and Phylogenetic Analysis

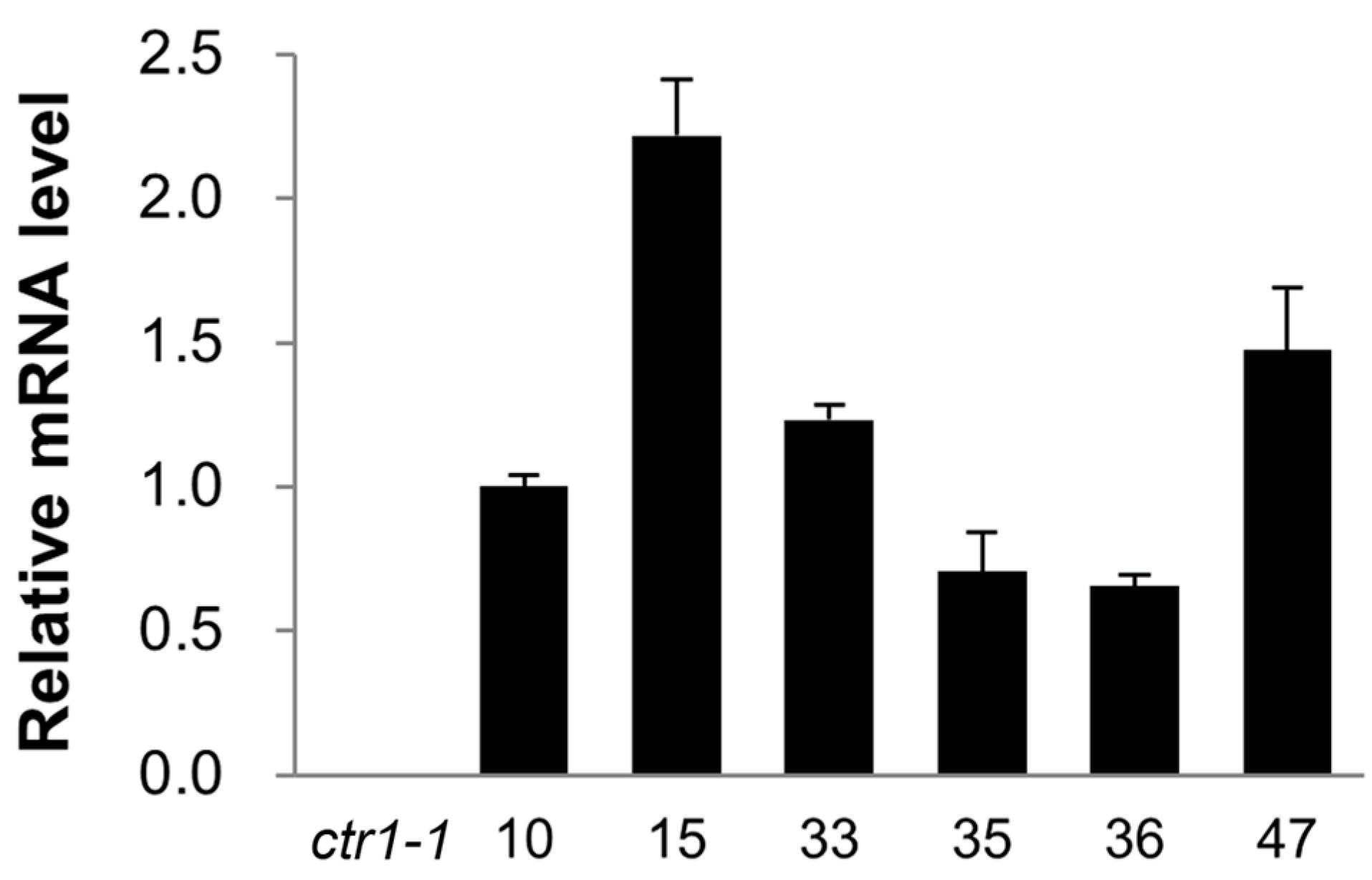
2.3. Ectopic Expression of CsCTR1 in an Arabidopsis ctr1-1 Mutant
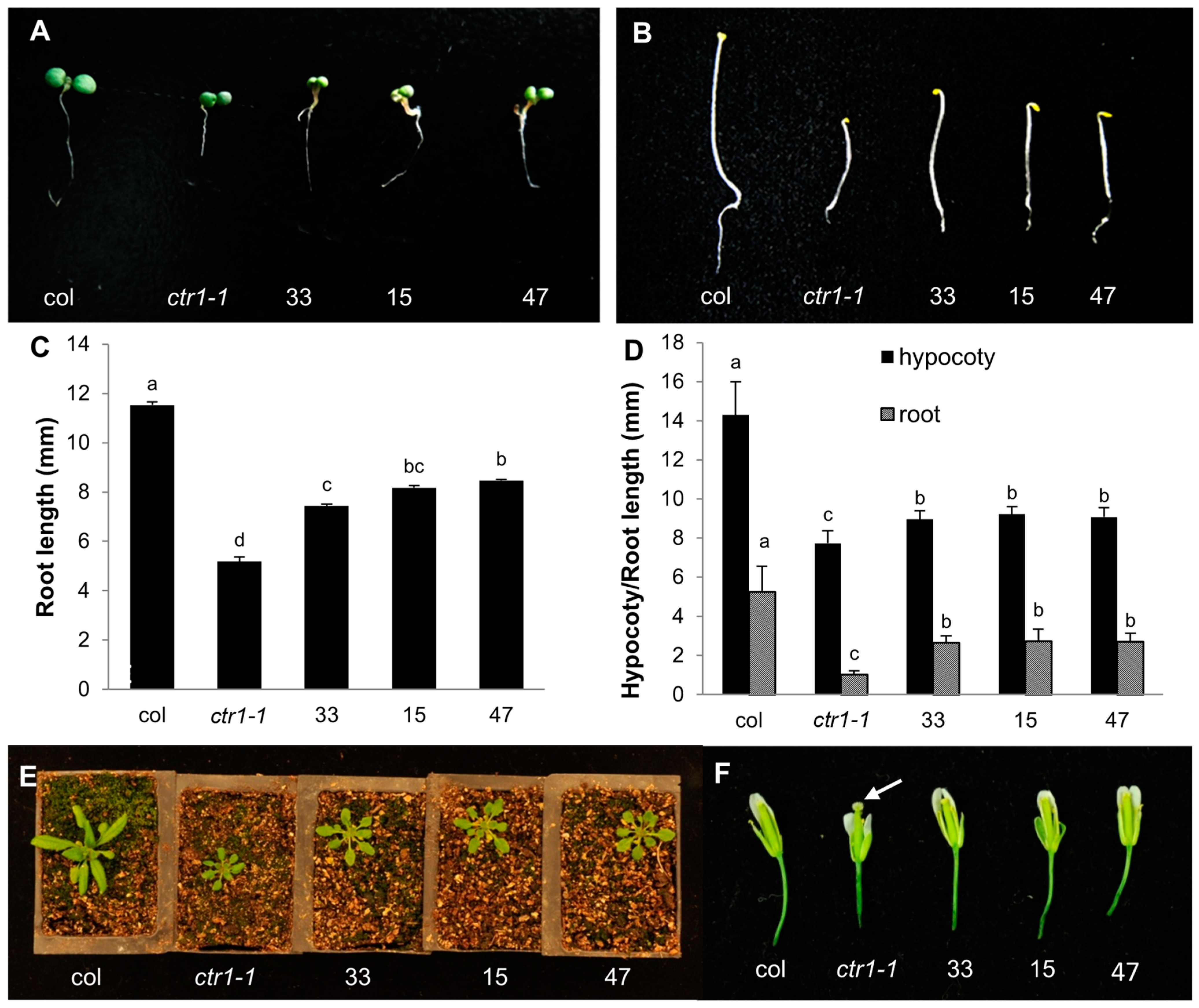
2.4. Partial Complementation of the Arabidopsis ctr1-1 Mutant by CsCTR1
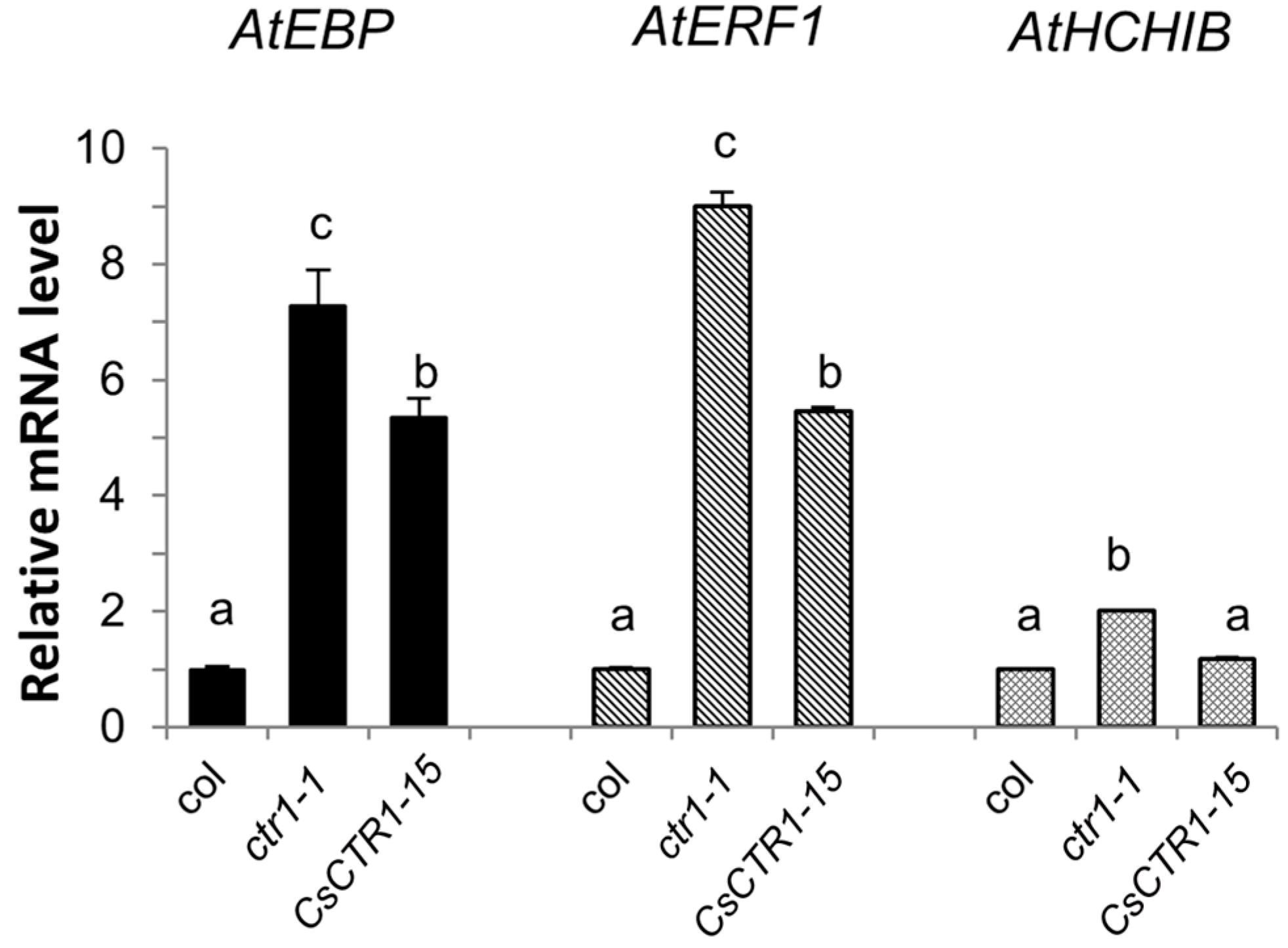
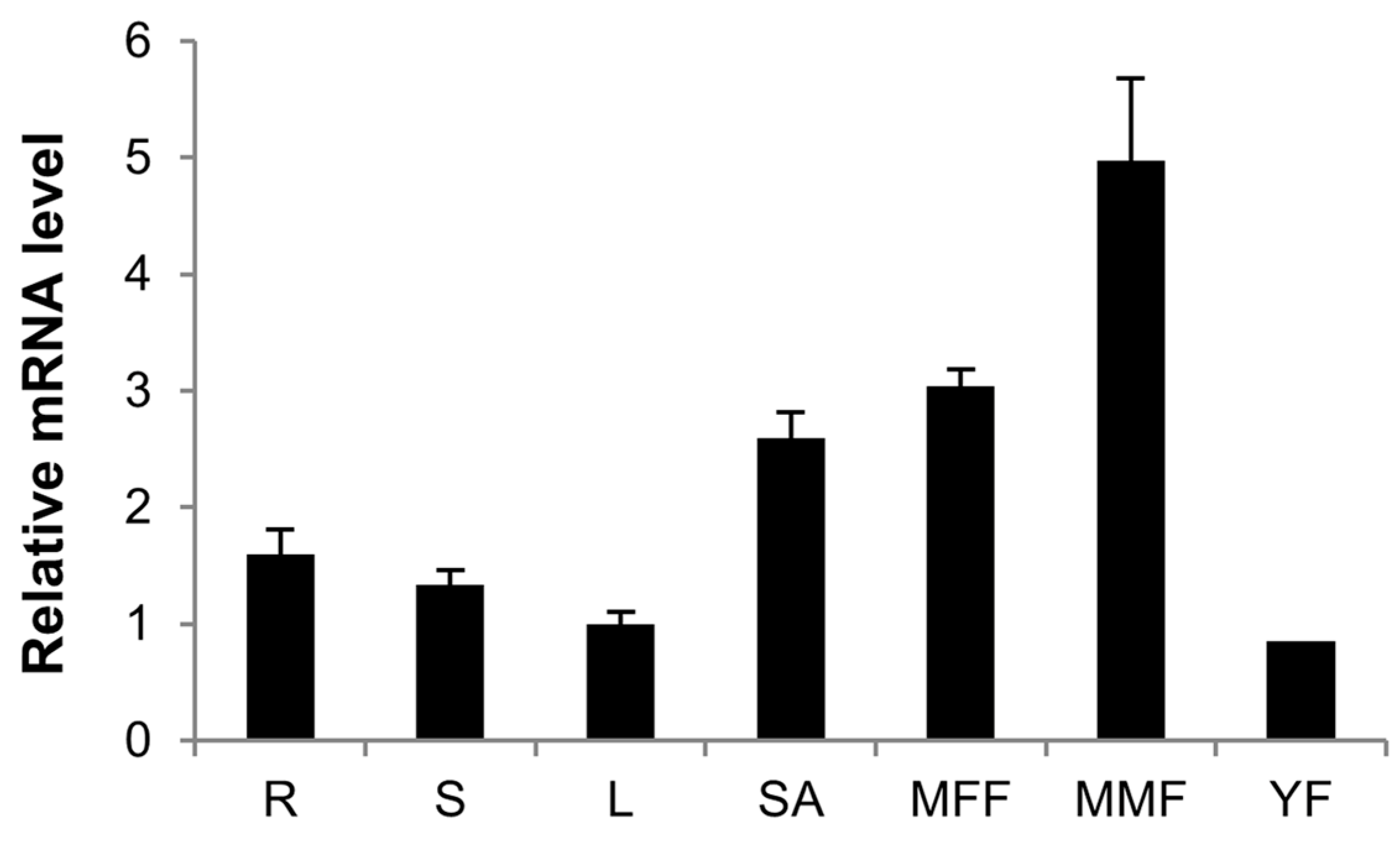
2.5. Expression Patterns of CsCTR1 in Different Organs
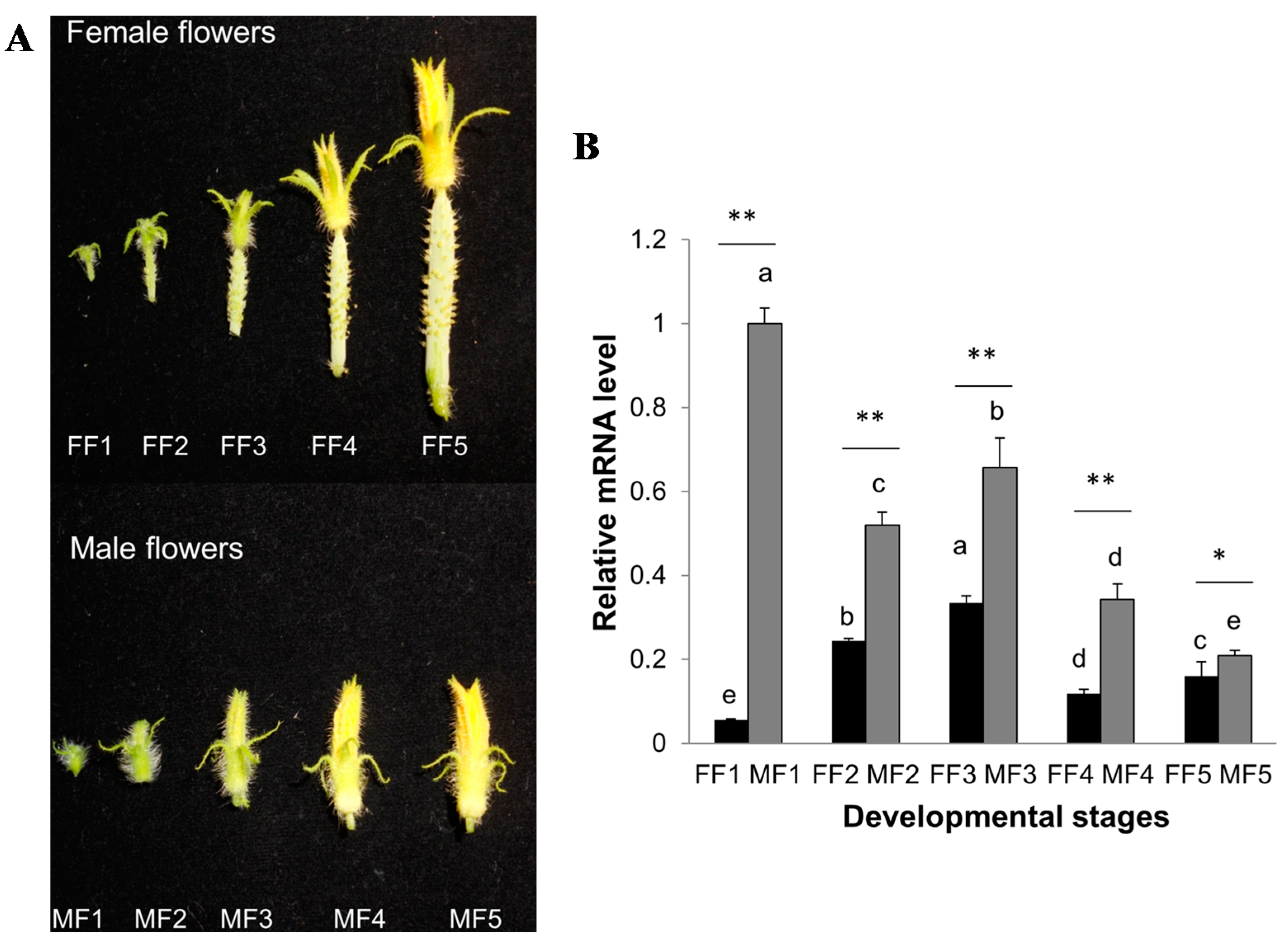
2.6. Expression Patterns of CsCTR1 during Flower Development
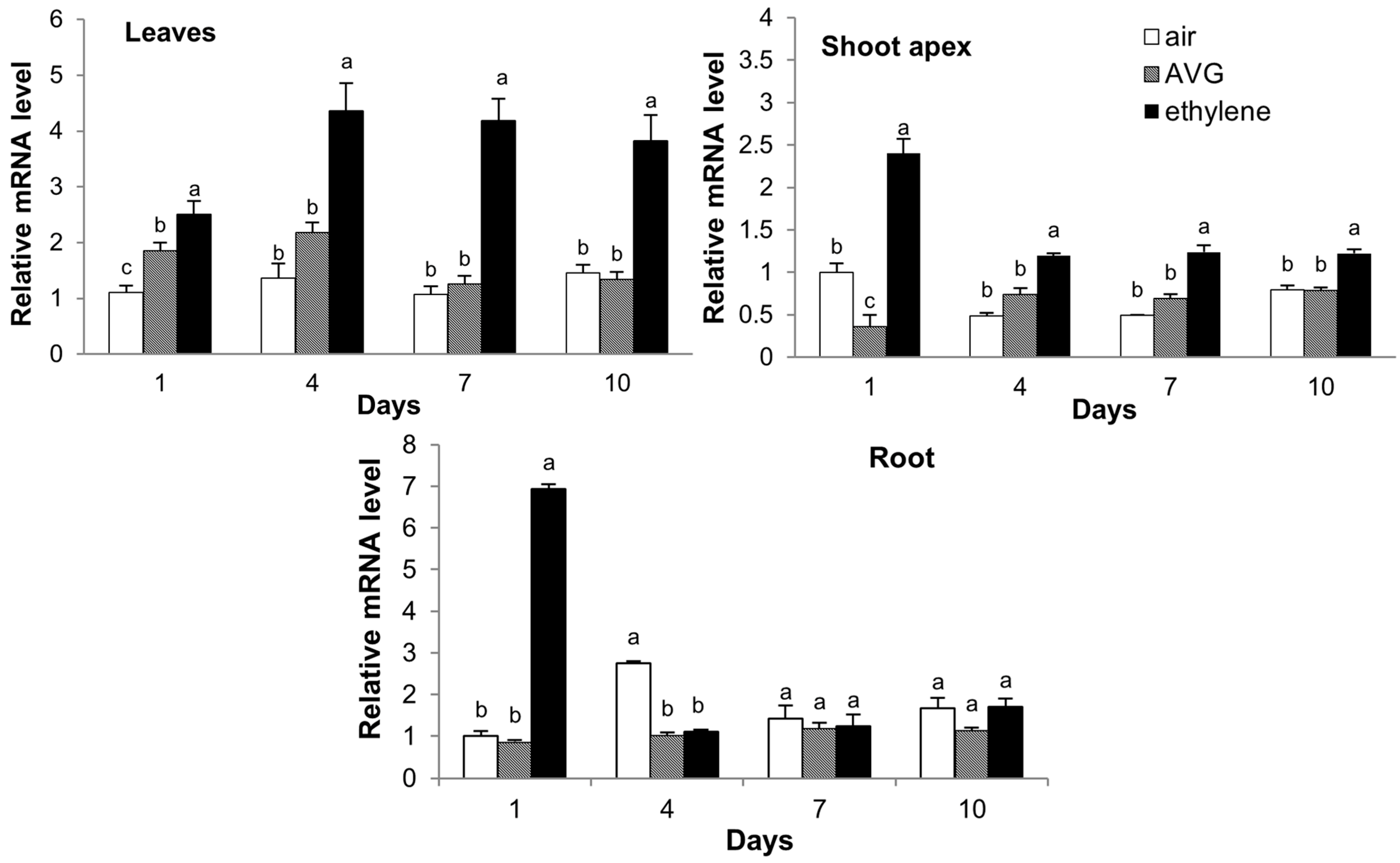
2.7. Expression of CsCTR1 in Response to Ethylene and 1-Aminoethoxyvinylglycine (AVG) Treatment
3. Discussion
4. Experimental Section
4.1. Plant Materials, Ethylene Treatments and Sampling
4.2. Cloning of the CsCTR1 Open Reading Frame (ORF)
4.3. Bioinformatic Analysis
4.4. Construction of Vector for Transformation
4.5. Construction of Vector for Transformation
4.6. Seedling Triple-Response Assay
4.7. Quantitative Real-Time PCR
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lin, Z.; Zhong, S.; Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 2009, 60, 3311–3336. [Google Scholar] [CrossRef]
- Guzman, P.; Ecker, J.R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 1990, 2, 513–523. [Google Scholar] [CrossRef]
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004, 7, 40–49. [Google Scholar] [CrossRef]
- Hua, J.; Chang, C.; Sun, Q.; Meyerowitz, E.M. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 1995, 269, 1712–1714. [Google Scholar]
- Hua, J.; Meyerowitz, E.M. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 1998, 94, 261–271. [Google Scholar] [CrossRef]
- Sakai, H.; Hua, J.; Chen, Q.G.; Chang, C.; Medrano, L.J.; Bleecker, A.B.; Meyerowitz, E.M. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 5812–5817. [Google Scholar]
- Dong, C.H.; Rivarola, M.; Resnick, J.S.; Maggin, B.D.; Chang, C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 2008, 53, 275–286. [Google Scholar]
- Huang, Y.; Li, H.; Hutchison, C.E.; Laskey, J.; Kieber, J.J. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 2003, 33, 221–233. [Google Scholar] [CrossRef]
- Kieber, J.J.; Rothenberg, M.; Roman, G.; Feldmann, K.A.; Ecker, J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 1993, 72, 427–441. [Google Scholar] [CrossRef]
- Wang, K.L.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, S131–S151. [Google Scholar]
- Clark, K.L.; Larsen, P.B.; Wang, X.; Chang, C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA 1998, 95, 5401–5406. [Google Scholar] [CrossRef]
- Ju, C.; Yoon, G.M.; Shemansky, J.M.; Lin, D.Y.; Ying, Z.I.; Chang, J.; Garrett, W.M.; Kessenbrock, M.; Groth, G.; Tucker, M.L.; et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 19486–19491. [Google Scholar]
- Qiao, H.; Shen, Z.; Huang, S.-S.C.; Schmitz, R.J.; Urich, M.A.; Briggs, S.P.; Ecker, J.R. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 2012, 338, 390–393. [Google Scholar] [CrossRef]
- Wen, X.; Zhang, C.; Ji, Y.; Zhao, Q.; He, W.; An, F.; Jiang, L.; Guo, H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012, 22, 1613–1616. [Google Scholar] [CrossRef]
- Ji, Y.; Guo, H. From endoplasmic reticulum (ER) to nucleus: EIN2 bridges the gap in ethylene signaling. Mol. Plant 2013, 6, 11–14. [Google Scholar] [CrossRef]
- Xu, A.; Zhang, W.; Wen, C.-K. Enhancing CTR1–10 ethylene response 2 is a novel allele involved in constitutive triple-response1-mediated ethylene receptor signaling in Arabidopsis. BMC Plant Biol. 2014, 14, 48. [Google Scholar] [CrossRef]
- Adams-Phillips, L.; Barry, C.; Kannan, P.; Leclercq, J.; Bouzayen, M.; Giovannoni, J. Evidence that CTR1-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Mol. Biol. 2004, 54, 387–404. [Google Scholar] [CrossRef]
- Leclercq, J.; Adams-Phillips, L.C.; Zegzouti, H.; Jones, B.; Latche, A.; Giovannoni, J.J.; Pech, J.C.; Bouzayen, M. LeCTR1, a tomato CTR1-like gene, demonstrates ethylene signaling ability in Arabidopsis and novel expression patterns in tomato. Plant Physiol. 2002, 130, 1132–1142. [Google Scholar] [CrossRef]
- Lin, Z.; Alexander, L.; Hackett, R.; Grierson, D. LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J. 2008, 54, 1083–1093. [Google Scholar] [CrossRef]
- Alexander, L.; Grierson, D. Ethylene biosynthesis and action in tomato: A model for climacteric fruit ripening. J. Exp. Bot. 2002, 53, 2039–2055. [Google Scholar] [CrossRef]
- Manzano, S.; Martinez, C.; Gomez, P.; Garrido, D.; Jamilena, M. Cloning and characterisation of two CTR1-like genes in Cucurbita pepo: Regulation of their expression during male and female flower development. Sex. Plant Reprod. 2010, 23, 301–313. [Google Scholar]
- Muller, R.; Owen, C.A.; Xue, Z.T.; Welander, M.; Stummann, B.M. Characterization of two CTR-like protein kinases in Rosa hybrida and their expression during flower senescence and in response to ethylene. J. Exp. Bot. 2002, 53, 1223–1225. [Google Scholar] [CrossRef]
- Yin, X.R.; Chen, K.S.; Allan, A.C.; Wu, R.M.; Zhang, B.; Lallu, N.; Ferguson, I.B. Ethylene-induced modulation of genes associated with the ethylene signalling pathway in ripening kiwifruit. J. Exp. Bot. 2008, 59, 2097–2108. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Jones, B.; Li, Z.G.; Lelievre, J.M.; Pech, J.C.; Latche, A. Isolation and characterization of four ethylene perception elements and their expression during ripening in pears (Pyrus communis L.) with/without cold requirement. J. Exp. Bot. 2003, 54, 1615–1625. [Google Scholar] [CrossRef]
- Kuroda, S.; Hirose, Y.; Shiraishi, M.; Davies, E.; Abe, S. Co-expression of an ethylene receptor gene, ERS1, and ethylene signaling regulator gene, CTR1, in Delphinium during abscission of florets. Plant Physiol. Biochem. 2004, 42, 745–751. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Kim, W.S.; el-Kereamy, A.; Jayasankar, S.; Svircev, A.M.; Brown, D.C. Isolation and characterization of four ethylene signal transduction elements in plums (Prunus salicina L.). J. Exp. Bot. 2007, 58, 3631–3643. [Google Scholar] [CrossRef]
- Wiersma, P.A.; Zhang, H.; Lu, C.; Quail, A.; Toivonen, P. Survey of the expression of genes for ethylene synthesis and perception during maturation and ripening of “sunrise” and “golden delicious” apple fruit. Postharvest Biol. Technol. 2007, 44, 204–211. [Google Scholar] [CrossRef]
- Rudich, J.; Halevy, A.H.; Kedar, N. The level of phytohormones in monoecious and gynoecious cucumbers as affected by photoperiod and ethephon. Plant Physiol. 1972, 50, 585–590. [Google Scholar] [CrossRef]
- Owens, K.; Peterson, C.; Tolla, G. Induction of perfect flowers on gynoecious muskmelon by silver nitrate and aminoethoxyvinylglycine. HortScience 1980, 15, 654–655. [Google Scholar]
- Yamasaki, S.; Fujii, N.; Matsuura, S.; Mizusawa, H.; Takahashi, H. The M locus and ethylene-controlled sex determination in andromonoecious cucumber plants. Plant Cell Physiol. 2001, 42, 608–619. [Google Scholar] [CrossRef]
- Knopf, R.R.; Trebitsh, T. The female-specific CS-ACS1G gene of cucumber. A case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched-chain amino acid transaminase gene. Plant Cell Physiol. 2006, 47, 1217–1228. [Google Scholar]
- Li, Z.; Huang, S.; Liu, S.; Pan, J.; Zhang, Z.; Tao, Q.; Shi, Q.; Jia, Z.; Zhang, W.; Chen, H.; et al. Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 2009, 182, 1381–1385. [Google Scholar] [CrossRef]
- Bie, B.; Pan, J.; He, H.; Yang, X.; Zhao, J.; Cai, R. Molecular cloning and expression analysis of the ethylene insensitive3 (EIN3) gene in cucumber (Cucumis sativus). Genet. Mol. Res. 2012, 12, 4179–4191. [Google Scholar]
- Yamasaki, S.; Fujii, N.; Takahashi, H. The ethylene-regulated expression of CS-ETR2 and CS-ERS genes in cucumber plants and their possible involvement with sex expression in flowers. Plant Cell Physiol. 2000, 41, 608–616. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Perl-Treves, R. Male to female conversion along the cucumber shoot: Approaches to studying sex genes and floral development in Cucumis sativus. In Sex Determination in Plants; Bios Scientific Publishers: Oxford, UK, 1999; pp. 189–215. [Google Scholar]
- Hu, H.L.; Do, Y.Y.; Huang, P.L. Expression profiles of a MhCTR1 gene in relation to banana fruit ripening. Plant Physiol. Biochem. 2012, 56, 47–55. [Google Scholar] [CrossRef]
- Gao, Z.; Chen, Y.F.; Randlett, M.D.; Zhao, X.C.; Findell, J.L.; Kieber, J.J.; Schaller, G.E. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 2003, 278, 34725–34732. [Google Scholar]
- Ma, N.; Tan, H.; Liu, X.; Xue, J.; Li, Y.; Gao, J. Transcriptional regulation of ethylene receptor and CTR genes involved in ethylene-induced flower opening in cut rose (Rosa hybrida) cv Samantha. J. Exp. Bot. 2006, 57, 2763–2773. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 2008, 55, 821–831. [Google Scholar]
- Han, Y.H.; Zhang, Z.H.; Liu, J.H.; Lu, J.Y.; Huang, S.W.; Jin, W.W. Distribution of the tandem repeat sequences and karyotyping in cucumber (Cucumis sativus L.) by fluorescence in situ hybridization. Cytogenet. Genome Res. 2008, 122, 80–88. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, Z.; Liu, J.; Staub, J.E.; Han, Y.; Cheng, Z.; Li, X.; Lu, J.; Miao, H.; Kang, H.; et al. An integrated genetic and cytogenetic map of the cucumber genome. PLoS One. 2009, 4, e5795. [Google Scholar] [CrossRef]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The clustal_x windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B., Jr.; Deerfield, D.W., II. GeneDoc: Analysis and visualization of genetic variation. EMBNEW. NEWS 1997, 4, 14. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bie, B.; Sun, J.; Pan, J.; He, H.; Cai, R. Ectopic Expression of CsCTR1, a Cucumber CTR-Like Gene, Attenuates Constitutive Ethylene Signaling in an Arabidopsis ctr1-1 Mutant and Expression Pattern Analysis of CsCTR1 in Cucumber (Cucumis sativus). Int. J. Mol. Sci. 2014, 15, 16331-16350. https://doi.org/10.3390/ijms150916331
Bie B, Sun J, Pan J, He H, Cai R. Ectopic Expression of CsCTR1, a Cucumber CTR-Like Gene, Attenuates Constitutive Ethylene Signaling in an Arabidopsis ctr1-1 Mutant and Expression Pattern Analysis of CsCTR1 in Cucumber (Cucumis sativus). International Journal of Molecular Sciences. 2014; 15(9):16331-16350. https://doi.org/10.3390/ijms150916331
Chicago/Turabian StyleBie, Beibei, Jin Sun, Junsong Pan, Huanle He, and Run Cai. 2014. "Ectopic Expression of CsCTR1, a Cucumber CTR-Like Gene, Attenuates Constitutive Ethylene Signaling in an Arabidopsis ctr1-1 Mutant and Expression Pattern Analysis of CsCTR1 in Cucumber (Cucumis sativus)" International Journal of Molecular Sciences 15, no. 9: 16331-16350. https://doi.org/10.3390/ijms150916331



