EPAS-1 Mediates SP-1-Dependent FBI-1 Expression and Regulates Tumor Cell Survival and Proliferation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Endothelial Pas Domain Protein-1 (EPAS-1) Up-Regulates the Expression Level of Factor Binding IST-1 (FBI-1)
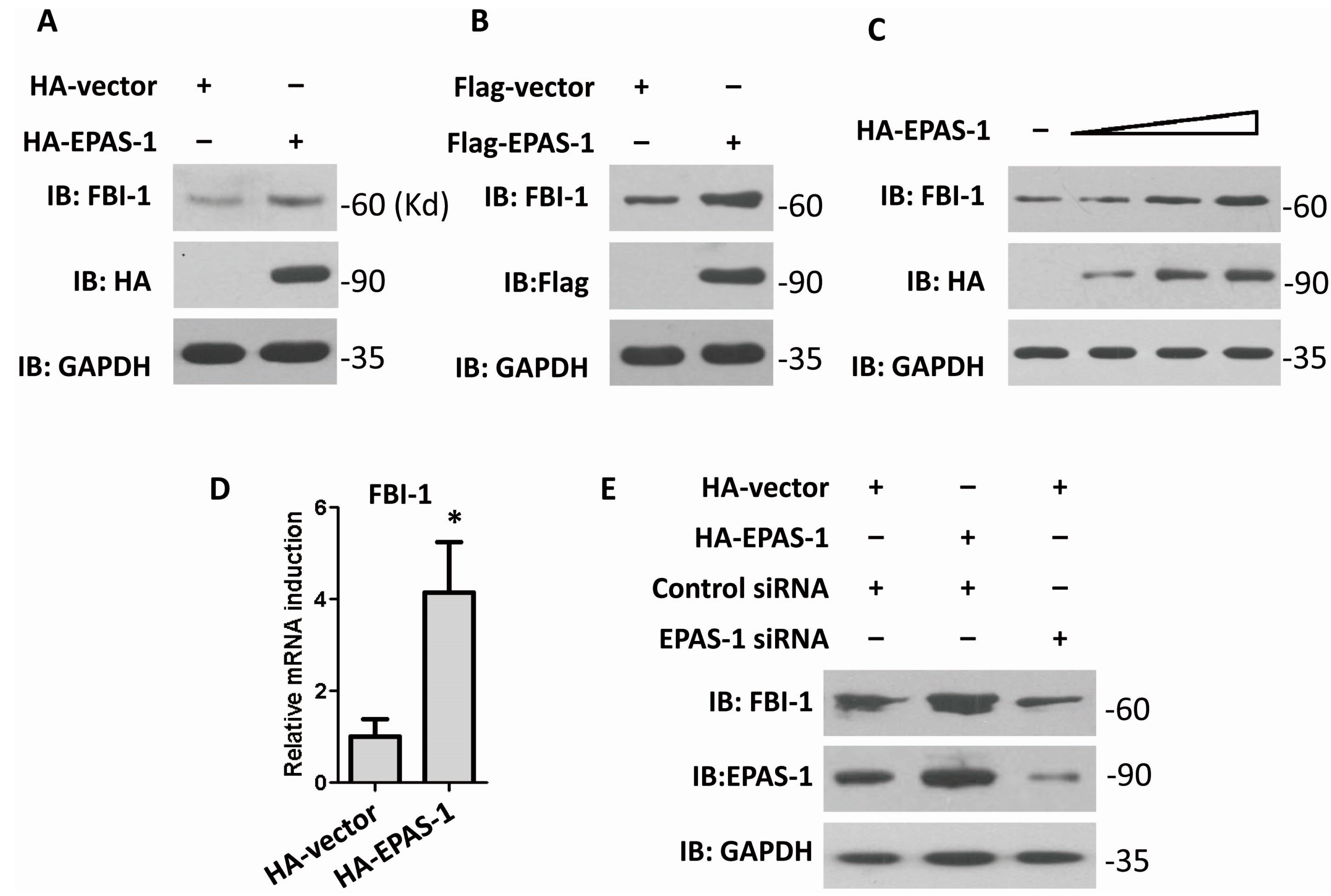
2.2. Both EPAS-1 and Specificity Protein-1 (SP-1) Control FBI-1 Expression Synergistically
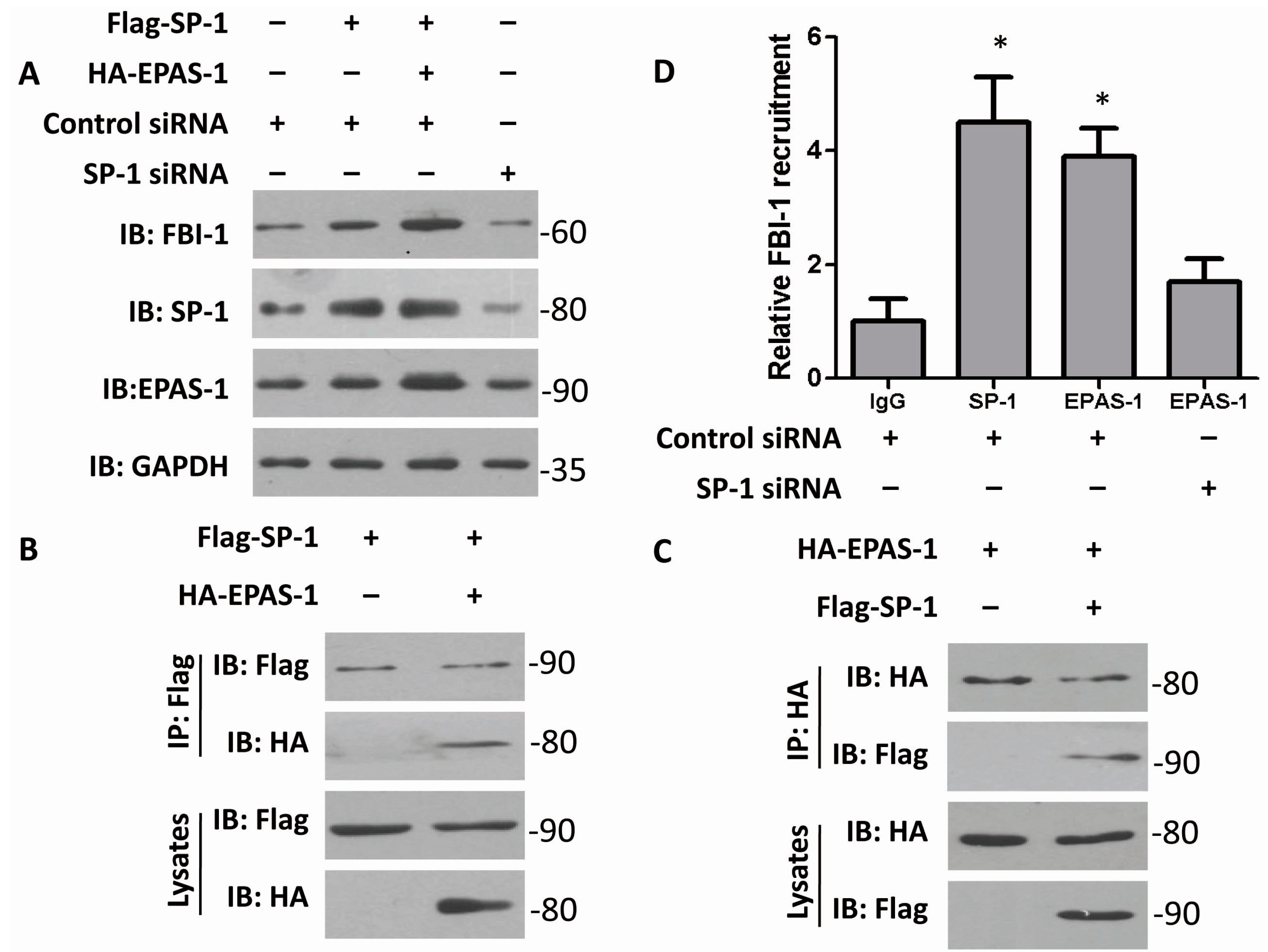
2.3. EPAS-1 Increases FBI-1-Luc Activity in a SP-1-Dependent Manner
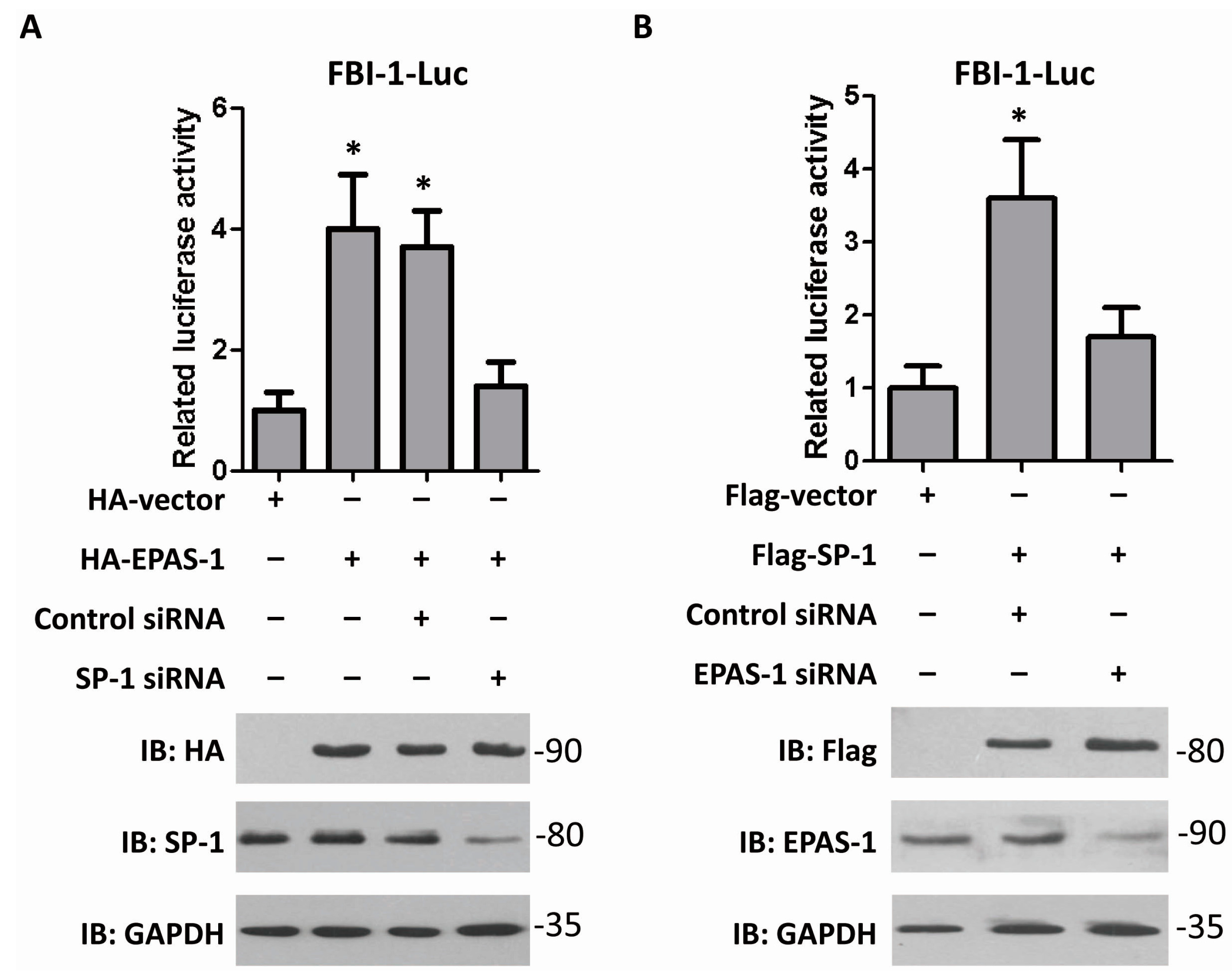
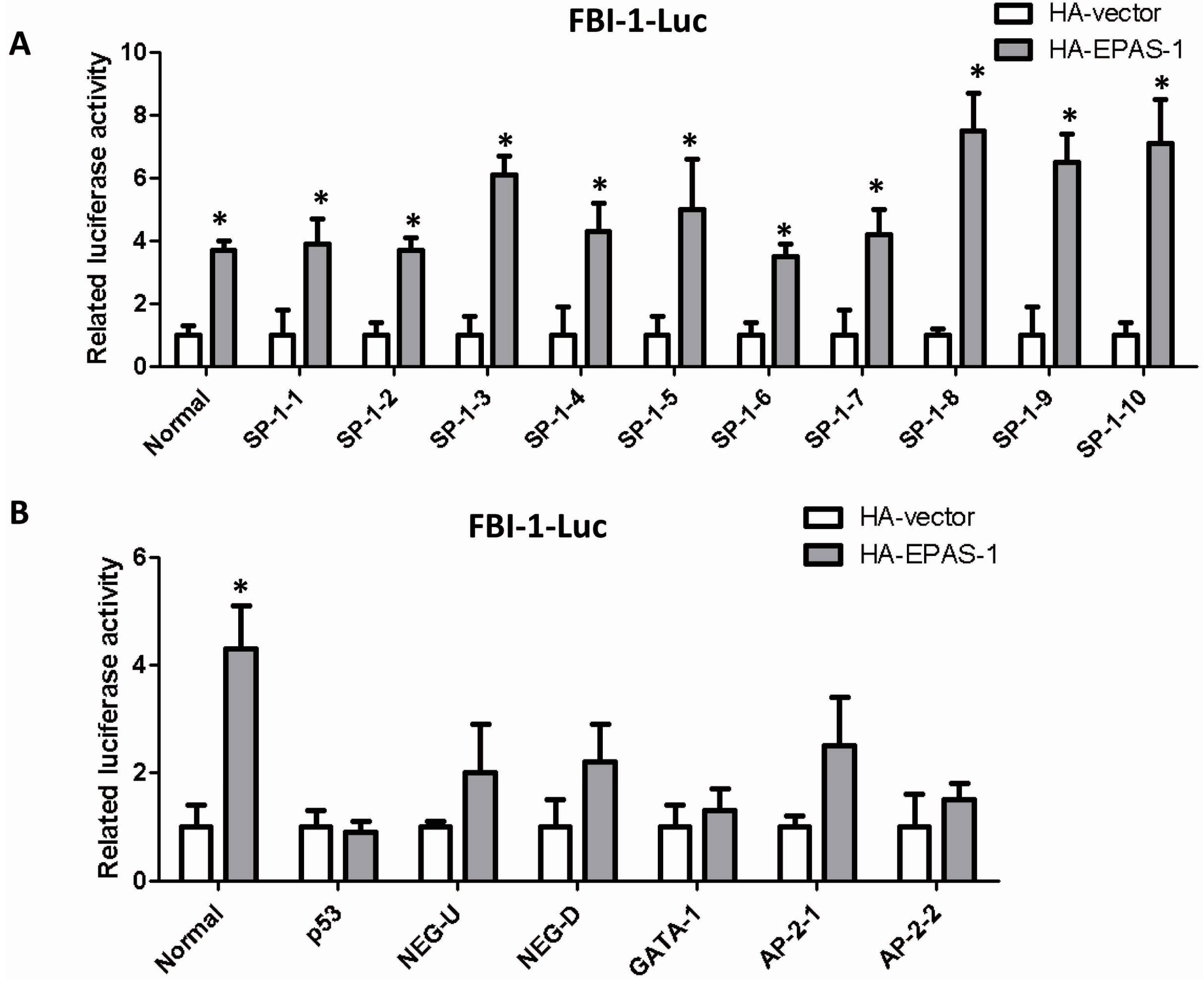
2.4. EPAS-1 Specifically Regulates SP-1-Mediated FBI-1 Expression
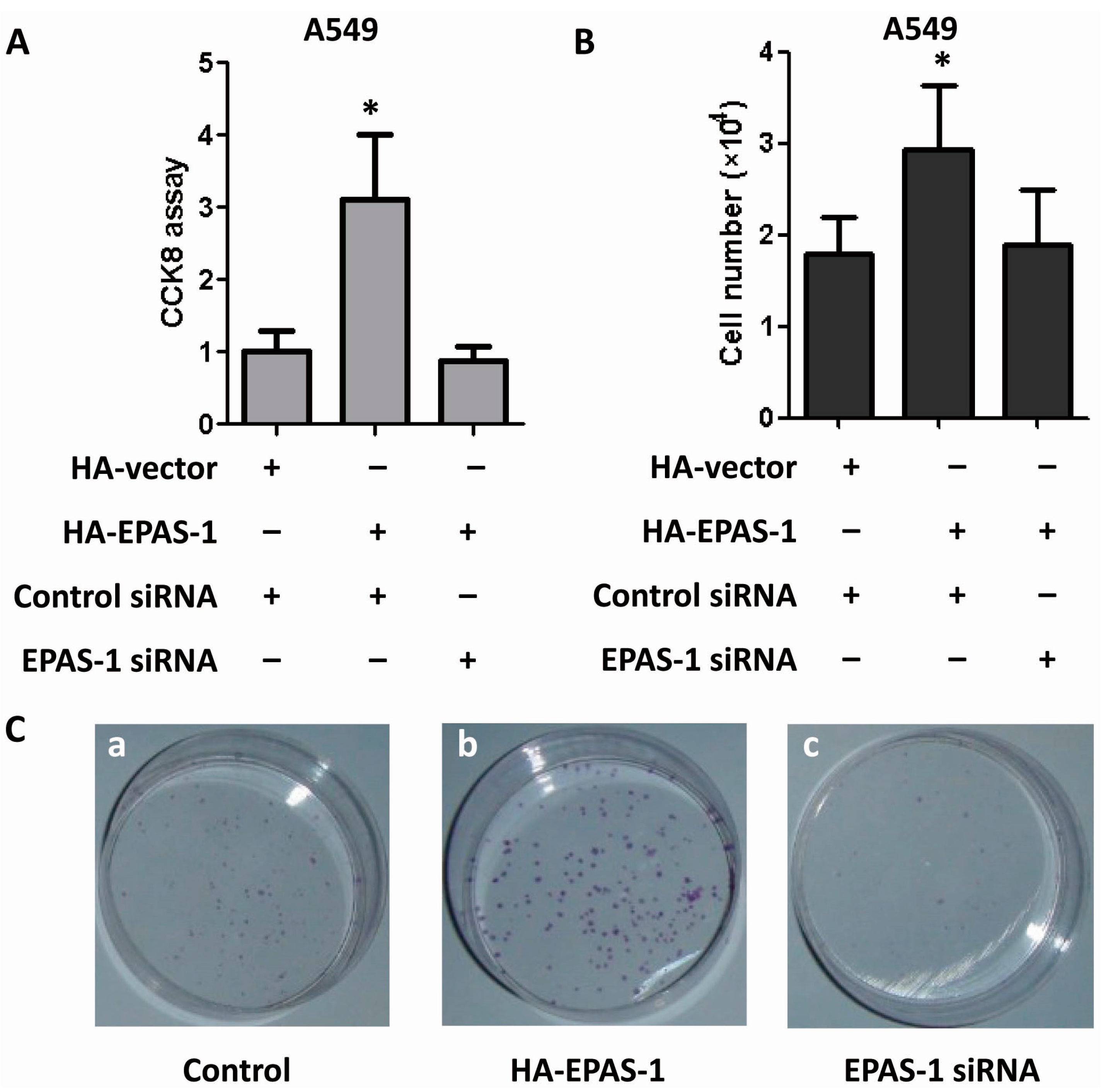
2.5. EPAS-1 Potentiates Human Lung Adenocarcinoma Cell Survival and Proliferation
3. Experimental Section
3.1. Plasmids and Antibodies
3.2. Cell Culture and Transfection
3.3. Reporter Gene Assay
3.4. RNA Interference
3.5. Co-Immunoprecipitation and Western Blot
3.6. CCK-8 Assay and Cell Counting
3.7. Clone Formation Assay
3.8. RNA Extraction and Quantitative Real-Time PCR
3.9. Chromatin Immunoprecipitation
3.10. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pessler, F.; Pendergrast, P.S.; Hernandez, N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol. Cell. Biol. 1997, 17, 3786–3798. [Google Scholar]
- Laudes, M.; Christodoulides, C.; Sewter, C.; Rochford, J.J.; Considine, R.V.; Sethi, J.K.; Vidal-Puig, A.; O’Rahilly, S. Role of the POZ zinc finger transcription factor FBI-1 in human and murine adipogenesis. J. Biol. Chem. 2004, 279, 11711–11718. [Google Scholar] [CrossRef]
- Liu, C.J.; Prazak, L.; Fajardo, M.; Yu, S.; Tyagi, N.; D., I.; Cesare, P.E. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J. Biol. Chem. 2004, 279, 47081–47091. [Google Scholar]
- Maeda, T.; Hobbs, R.M.; Pandolfi, P.P. The transcription factor Pokemon: A new key player in cancer pathogenesis. Cancer Res. 2005, 65, 8575–8578. [Google Scholar] [CrossRef]
- Widom, R.L.; Lee, J.Y.; Joseph, C.; Gordon-Froome, I.; Korn, J.H. The hcKrox gene family regulates multiple extracellular matrix genes. Matrix Biol. 2001, 20, 451–462. [Google Scholar] [CrossRef]
- Pessler, F.; Hernandez, N. Flexible DNA binding of the BTB/POZ-domain protein FBI-1. J. Biol. Chem. 2003, 278, 29327–29335. [Google Scholar] [CrossRef]
- Maeda, T.; Hobbs1, R.M.; Merghoub1, T.; Guernah1, I.; Zelent, A.; Cordon-Cardo, C.; Teruya-Feldstein, J.; Pandolfi1, P.P. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature 2005, 433, 278–285. [Google Scholar] [CrossRef]
- Lin, C.C.; Zhou, J.P.; Liu, Y.P.; Liu, J.J.; Yan, X.N.; Amarsanaa, J.; Zhang, Z.P.; Guleng, B.; Ren, J.L. The silencing of Pokemon attenuates the proliferation of hepatocellular carcinoma cells in vitro and in vivo by inhibiting the PI3K/Akt pathway. PLoS One 2012, 7, e51916. [Google Scholar]
- Cui, J.; Yang, Y.; Zhang, C.; Hu, P.; Kan, W.; Bai, X.; Liu, X.; Song, H. FBI-1 functions as a novel AR co-repressor in prostate cancer cells. Cell. Mol. Life Sci. 2011, 68, 1091–1103. [Google Scholar] [CrossRef]
- Jeon, B.N.; Yoo, J.Y.; Choi, W.I.; Lee, C.E.; Yoon, H.G.; Hur, M.W. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses transcription of the tumor suppressor Rb gene via binding competition with Sp1 and recruitment of co-repressors. J. Biol. Chem. 2008, 283, 33199–33210. [Google Scholar] [CrossRef]
- Keith, B.; Simon, M.C. Hypoxia-inducible factors, stem cells, and cancer. Cell 2007, 129, 465–472. [Google Scholar] [CrossRef]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef]
- Koizume, S.; Ito, S.; Miyagi, E.; Hirahara, F.; Nakamura, Y.; Sakuma, Y.; Osaka, H.; Takano, Y.; Ruf, W.; Miyagi, Y. HIF2α-Sp1 interaction mediates a deacetylation-dependent FVII-gene activation under hypoxic conditions in ovarian cancer cells. Nucleic Acids Res. 2012, 40, 5389–5401. [Google Scholar] [CrossRef]
- Gidoni, D.; Dynan, W.S.; Tjian, R. Multiple specific contacts between a mammalian transcription factor and its cognate promoters. Nature 1984, 312, 409–413. [Google Scholar] [CrossRef]
- Choi, W.I.; Jeon, B.N.; Yun, C.O.; Kim, P.H.; Kim, S.E.; Choi, K.Y.; Kim, S.H.; Hur, M.W. Proto-oncogene FBI-1 represses transcription of p21CIP1 by inhibition of transcription activation by p53 and Sp1. J. Biol. Chem. 2009, 284, 12633–12644. [Google Scholar]
- Suske, G. The Sp-family of transcription factors. Gene 1999, 238, 291–300. [Google Scholar] [CrossRef]
- Cui, J.; Meng, X.; Gao, X.; Tan, G. Curcumin decreases the expression of Pokemon by suppressing the binding activity of the Sp1 protein in human lung cancer cells. Mol. Biol. Rep. 2010, 37, 1627–1632. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, X.; Zhu, X.; Zhang, C.; Yang, Z.; Xu, L.; Huang, P. Cloning and functional analysis of 5'-upstream region of the Pokemon gene. FEBS J. 2008, 275, 1860–1873. [Google Scholar] [CrossRef]
- Zhang, P.; Ma, X.; Song, E.; Chen, W.H.; Pang, H.G.; Ni, D.; Gao, Y.; Fan, Y.; Ding, Q.; Zhang, Y.; Zhang, X. Tubulin cofactor A functions as a novel positive regulator of ccRCC progression, invasion and metastasis. Int. J. Cancer 2013, 133, 2801–2811. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, X.; Cao, P.; Li, Z.; Wu, D.; Wang, X.; Liang, G. EPAS-1 Mediates SP-1-Dependent FBI-1 Expression and Regulates Tumor Cell Survival and Proliferation. Int. J. Mol. Sci. 2014, 15, 15689-15699. https://doi.org/10.3390/ijms150915689
Wang X, Cao P, Li Z, Wu D, Wang X, Liang G. EPAS-1 Mediates SP-1-Dependent FBI-1 Expression and Regulates Tumor Cell Survival and Proliferation. International Journal of Molecular Sciences. 2014; 15(9):15689-15699. https://doi.org/10.3390/ijms150915689
Chicago/Turabian StyleWang, Xiaogang, Peng Cao, Zhiqing Li, Dongyang Wu, Xi Wang, and Guobiao Liang. 2014. "EPAS-1 Mediates SP-1-Dependent FBI-1 Expression and Regulates Tumor Cell Survival and Proliferation" International Journal of Molecular Sciences 15, no. 9: 15689-15699. https://doi.org/10.3390/ijms150915689



