ZmCIPK21, A Maize CBL-Interacting Kinase, Enhances Salt Stress Tolerance in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
2.1. Cloning and Phylogenetic Analysis of the ZmCIPK21 Gene
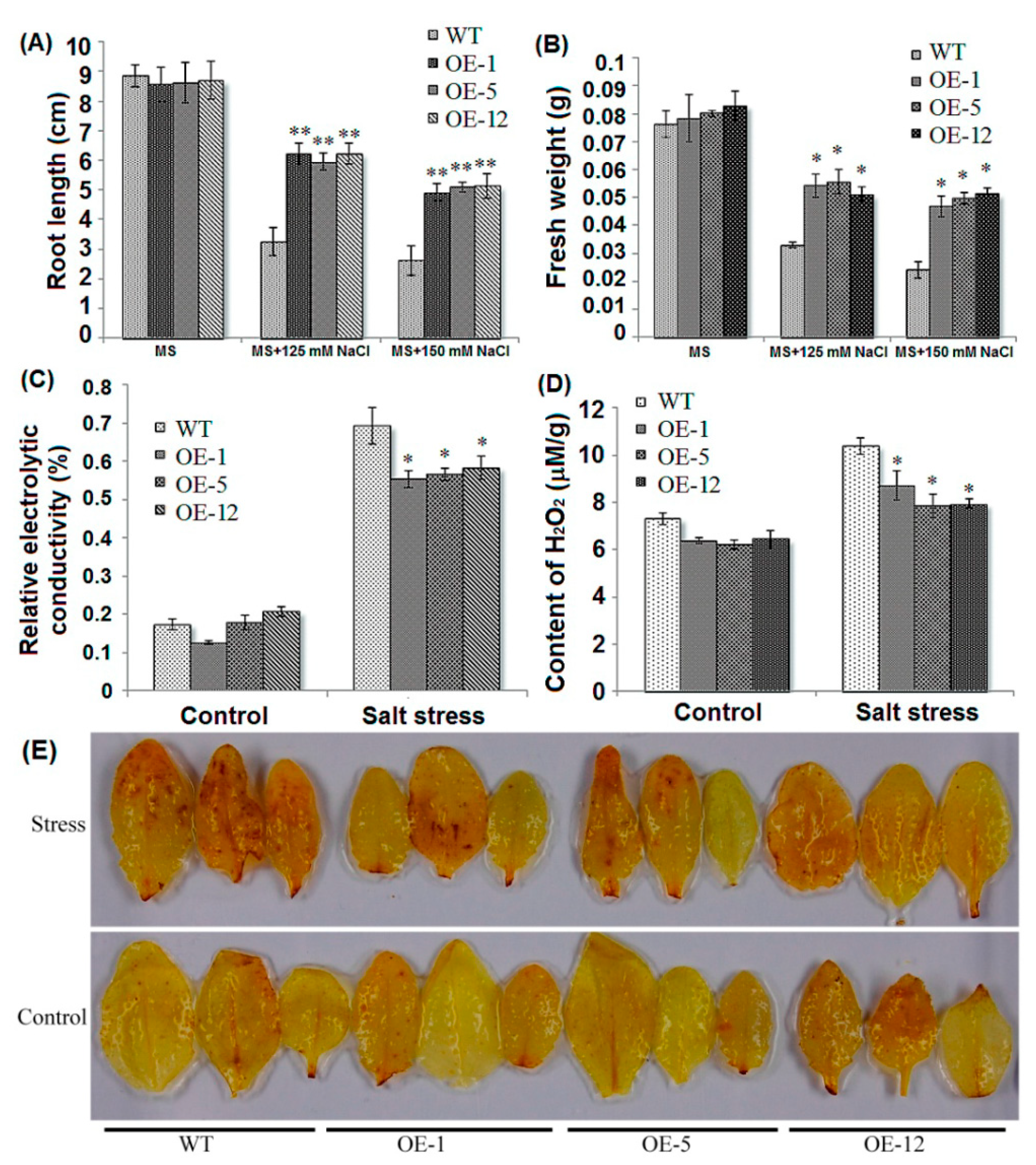
2.2. Over-Expression of ZmCIPK21 Enhanced Salt Tolerance in Arabidopsis
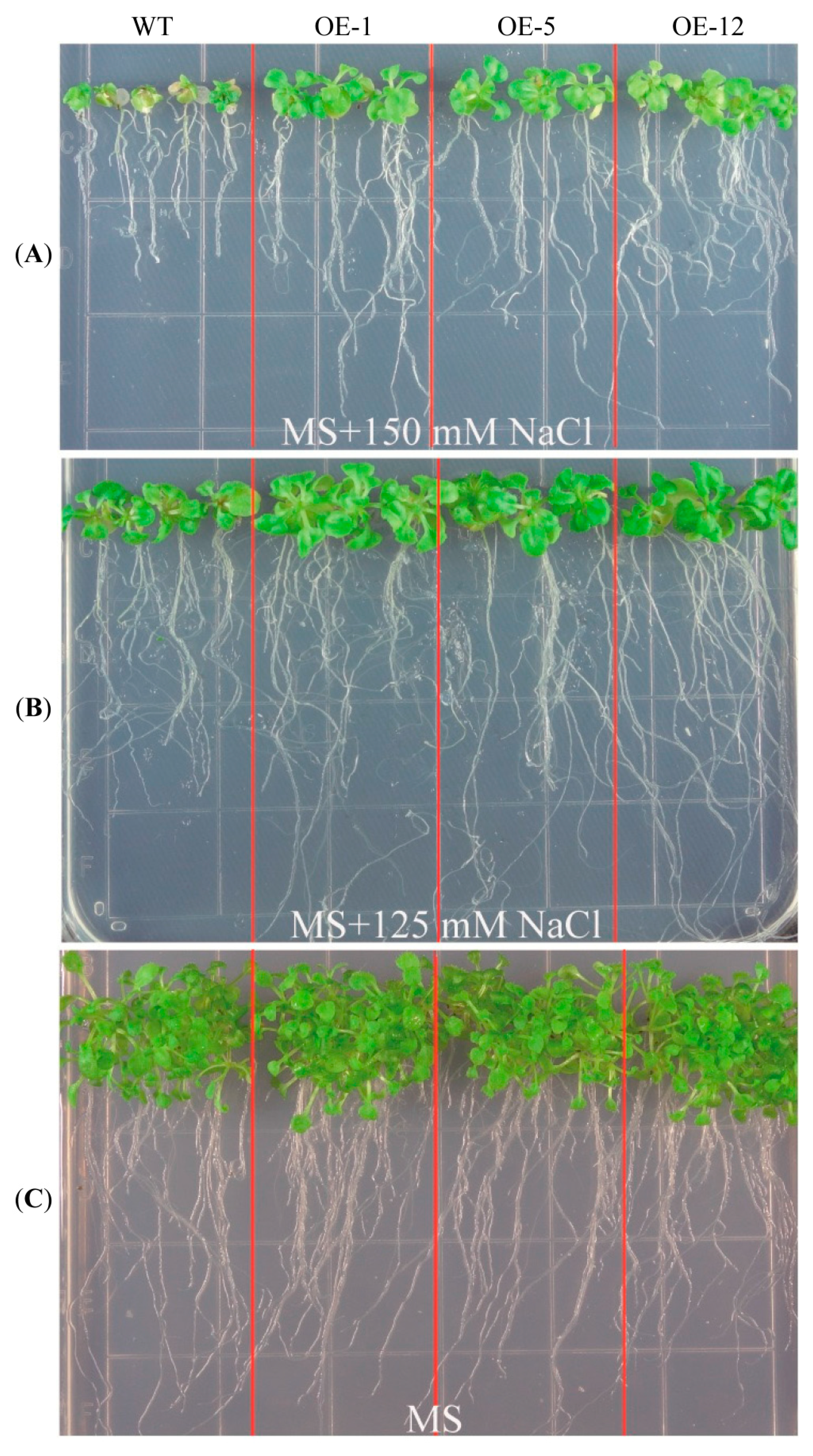
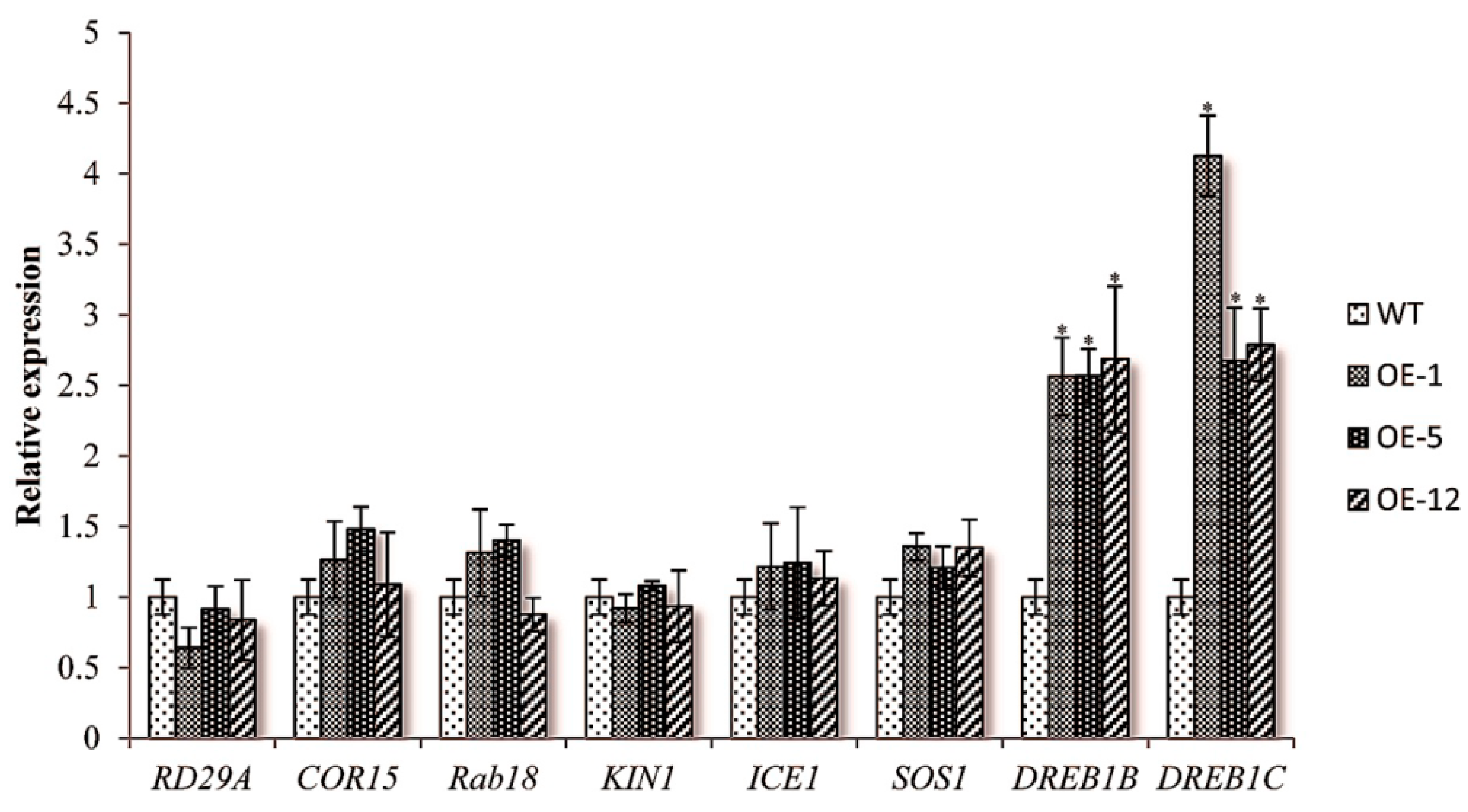
2.3. Over-Expressing of ZmCIPK21 Resulted in Up-Regulation of Two DREB Genes in Arabidopsis
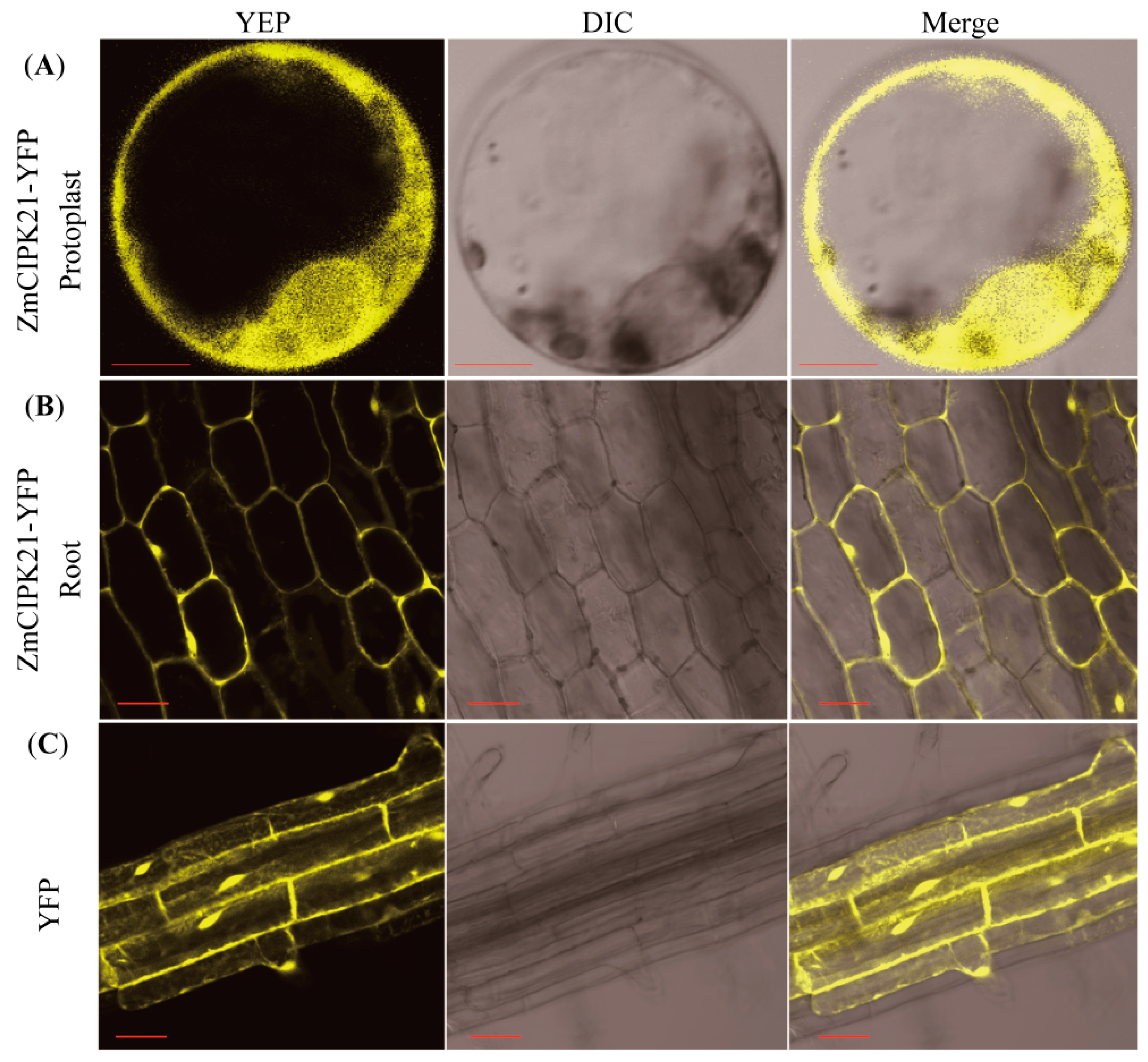
2.4. Subcellular Localization of ZmCIPK21
2.5. ZmCIPK21 Rescued the Sensitivity of the Arabidopsis Cipk1-2 Mutant
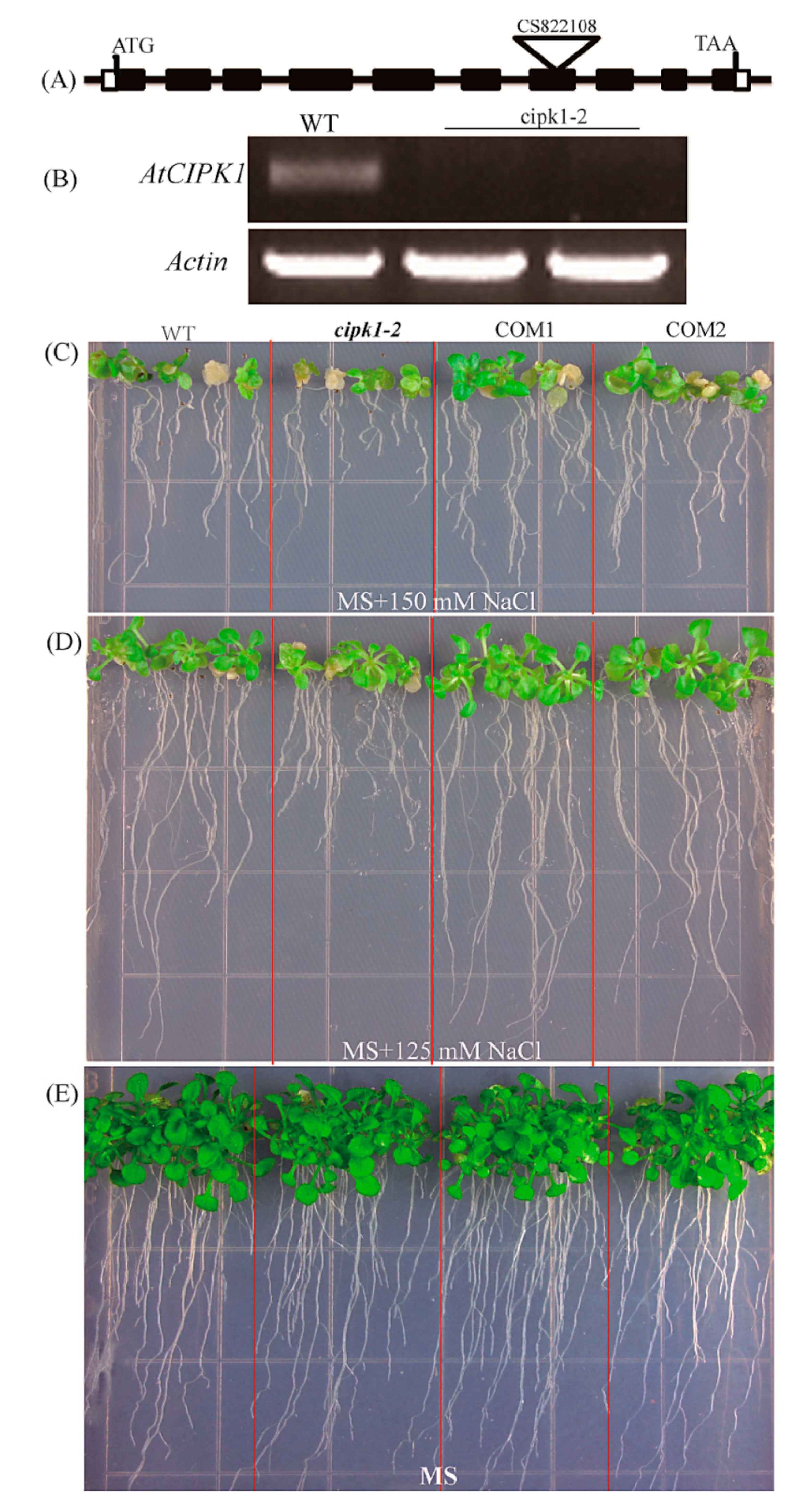
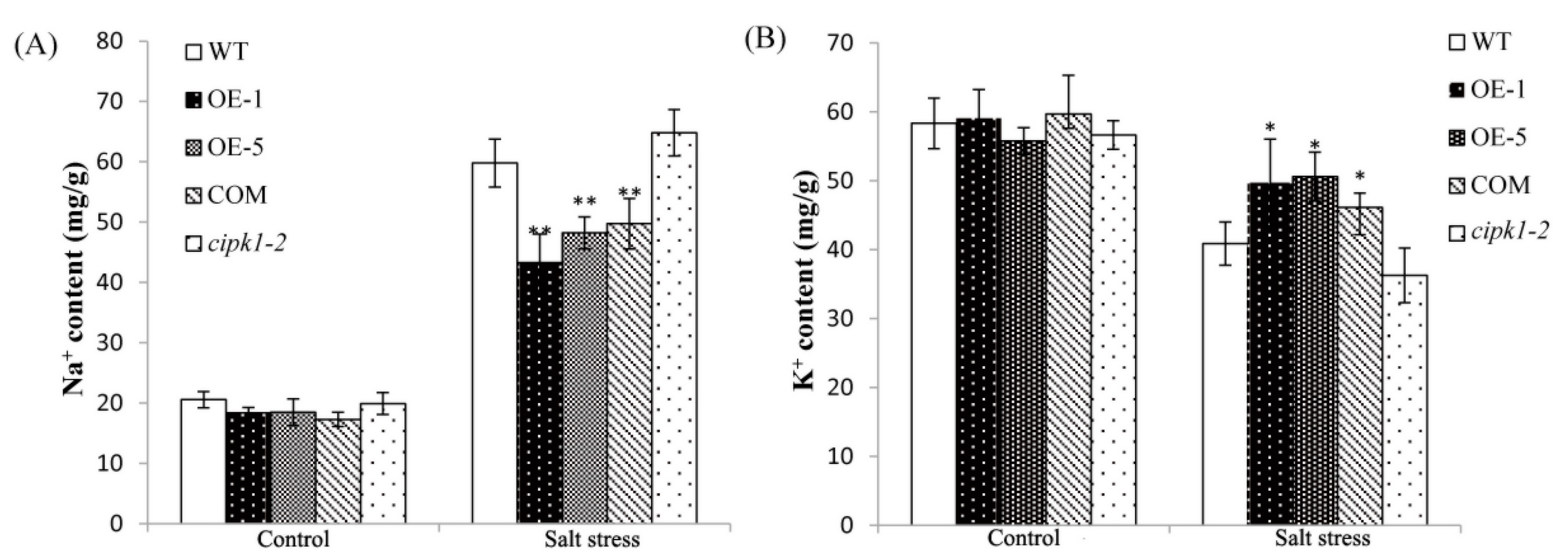
2.6. Over-Expressing of ZmCIPK21 Maintained Na+ and K+ Homeostasis in Plant Cells
3. Discussion
4. Experimental Section
4.1. Arabidopsis Growth and Analysis of Salt Tolerance
4.2. Generation of Transgenic Plants Over-Expressing the ZmCIPK21 Gene
4.3. Quantitative PCR Analysis of Gene Expression in Arabidopsis
| Gene | Upstream Primer (5'-3') | Downstream Primer (5'-3') |
|---|---|---|
| RD29A | CCCGGATCCTTTTCTGATATGGTTGCC | GCCCTCGAGCCGAACAATTTATTAACC |
| COR15 | AACTCTGCCGCCTTGTTTGC | CTTGGTGCAAGTGCTGTGAT |
| Rab18 | AGCAGCTTGCTTCTCAGCTT | ATTCCTTCTTCCCGGCTAACCT |
| KIN1 | TGCCTTCCAAGCCGGTCAGA | AGGCCGGTCTTGTCCTTCAC |
| ICE1 | GGAGATGCAATTGATTATCTGAAG | AGTCAGGATCCGATCAGATCATACC |
| SOS1 | GACGGGAGAATCAATCGAAA | TGCTCTTGCTCTCGTCTCAA |
| DREB1B | GGCGTTGGCTTTTCAAGATG | AAGTCGGCATCCCAAACATT |
| DREB1C | TTCGATTTTTATTTCCATTTTTGG | CCAAACGTCCTTGAGTCTTGAT |
| Actin | TAACCCAAAGGCAACAGAG | CTTGGTGCAAGTGCTGTGAT |
| ZmCIPK21 | CAGAGGGTGGCAAGAAGGAATTGT | GTTGCTTCAGTCCATTTCCFGTACC |
| ZmGAPDH | GGGTGAGGCTGGTGCTGAGTATGT | TTAGCAAGGGGAGCAAGGCAGTT |
4.4. Subcellular Localization
4.5. Measurement of Na+ and K+ Content
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yang, Q.; Chen, Z.-Z.; Zhou, X.-F.; Yin, H.-B.; Li, X.; Xin, X.-F.; Hong, X.-H.; Zhu, J.-K.; Gong, Z. Over-expressing of SOS (salt overly sensitive) genes increases salt tolerance in transgenic arabidopsis. Mol. Plant 2009, 2, 22–31. [Google Scholar] [CrossRef]
- Hasegawa, K.; Li, Y. Explosion investigation of asphalt-salt mixtures in a reprocessing plant. J. Hazard. Mater. 2000, 79, 241–267. [Google Scholar] [CrossRef]
- Shi, H.; Xiong, L.; Stevenson, B.; Lu, T.; Zhu, J.K. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 2002, 14, 575–588. [Google Scholar] [CrossRef]
- Kim, M.J.; Shin, R.; Schachtman, D.P. A nuclear factor regulates abscisic acid responses in arabidopsis. Plant Physiol. 2009, 151, 1433–1445. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
- Lloret, F.; Lobo, A.; Estevan, H.; Maisongrande, P.; Vayreda, J.; Terradas, J. Woody plant richness and NDVI response to drought events in catalonian (northeastern spain) forests. Ecology 2007, 88, 2270–2279. [Google Scholar] [CrossRef]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef]
- D’Angelo, C.; Weinl, S.; Batistic, O.; Pandey, G.K.; Cheong, Y.H.; Schültke, S.; Albrecht, V.; Ehlert, B.; Schulz, B.; Harter, K.; et al. Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 2006, 48, 857–872. [Google Scholar] [CrossRef]
- Xiong, L.; Wang, R.G.; Mao, G.; Koczan, J.M. Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol. 2006, 142, 1065–1074. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. 2006, 27, 141–177. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of drebs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef]
- Li, D.; Zhang, Y.; Hu, X.; Shen, X.; Ma, L.; Su, Z.; Wang, T.; Dong, J. Transcriptional profiling of medicago truncatula under salt stress identified a novel CBF transcription factor MTCBF4 that plays an important role in abiotic stress responses. BMC Plant Biol. 2011, 11, 109. [Google Scholar] [CrossRef]
- Chen, X.; Gu, Z.; Xin, D.; Hao, L.; Liu, C.; Huang, J.; Ma, B.; Zhang, H. Identification and characterization of putative cipk genes in maize. J. Genet. Genomics 2011, 38, 77–87. [Google Scholar] [CrossRef]
- Yu, Y.; Xia, X.; Yin, W.; Zhang, H. Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul. 2007, 52, 101–110. [Google Scholar] [CrossRef]
- Chen, X.L.; Li, J.P.; Hao, X.Y.; Chen, G.; Huang, Q.S. Cloning and characterization of a maize ZMCIPK21 gene. J. Nucl. Agric. Sci. 2012, 26, 862–867. [Google Scholar]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yanagawa, K.; Takasaki, M.; Inazu, M.; Takeda, H.; Matsumiya, T. Changes of the redox dynamics of α-tocopherol in erythrocyte membranes and plasma in streptozocin induced rats. Nihon Ronen Igakkai Zasshi 2000, 37, 611–612. [Google Scholar] [CrossRef]
- Pandey, G.K.; Cheong, Y.H.; Kim, K.N.; Grant, J.J.; Li, L.; Hung, W.; DʼAngelo, C.; Weinl, S.; Kudla, J.; Luan, S. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in arabidopsis. Plant Cell 2004, 16, 1912–1924. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Yu, Q.; An, L.; Li, W. The CBL–CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014, 33, 203–214. [Google Scholar] [CrossRef]
- Piao, H.L.; Xuan, Y.H.; Park, S.H.; Je, B.I.; Park, S.J.; Kim, C.M.; Huang, J.; Wang, G.K.; Kim, M.J.; Kang, S.M.; et al. Oscipk31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol. Cells 2010, 30, 19–27. [Google Scholar] [CrossRef]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef]
- Zhu, Y.; Ye, J.; Huizinga, J.D. Clotrimazole-sensitive k+ currents regulate pacemaker activity in interstitial cells of cajal. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G1715–G1725. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef]
- Qi, Z.; Spalding, E.P. Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+–H+ antiporter during salinity stress. Plant Physiol. 2004, 136, 2548–2555. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef]
- Deng, X.; Zhou, S.; Hu, W.; Feng, J.; Zhang, F.; Chen, L.; Huang, C.; Luo, Q.; He, Y.; Yang, G.; et al. Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol. Plant 2013, 149, 367–377. [Google Scholar]
- Gay, C.; Gebicki, J.M. A critical evaluation of the effect of sorbitol on the ferric-xylenol orange hydroperoxide assay. Anal. Biochem. 2000, 284, 217–220. [Google Scholar] [CrossRef]
- Karimi, M.; Inze, D.; Depicker, A. Gateway vectors for agrobacterium-mediated plant transformation. Trends Plant. Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006, 45, 616–629. [Google Scholar] [CrossRef]
- Garg, A.K.; Kim, J.K.; Owens, T.G.; Ranwala, A.P.; Choi, Y.D.; Kochian, L.V.; Wu, R.J. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chen, X.; Huang, Q.; Zhang, F.; Wang, B.; Wang, J.; Zheng, J. ZmCIPK21, A Maize CBL-Interacting Kinase, Enhances Salt Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2014, 15, 14819-14834. https://doi.org/10.3390/ijms150814819
Chen X, Huang Q, Zhang F, Wang B, Wang J, Zheng J. ZmCIPK21, A Maize CBL-Interacting Kinase, Enhances Salt Stress Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences. 2014; 15(8):14819-14834. https://doi.org/10.3390/ijms150814819
Chicago/Turabian StyleChen, Xunji, Quansheng Huang, Fan Zhang, Bo Wang, Jianhua Wang, and Jun Zheng. 2014. "ZmCIPK21, A Maize CBL-Interacting Kinase, Enhances Salt Stress Tolerance in Arabidopsis thaliana" International Journal of Molecular Sciences 15, no. 8: 14819-14834. https://doi.org/10.3390/ijms150814819




