Genetic Variants of APOC3 Promoter and HLA-B Genes in an HIV Infected Cohort in Northern South Africa: A Pilot Study
Abstract
:1. Introduction
2. Results
2.1. Study Population and Clinical Data
2.2. APOC3 Promoter Region Amplification and RFLP Results
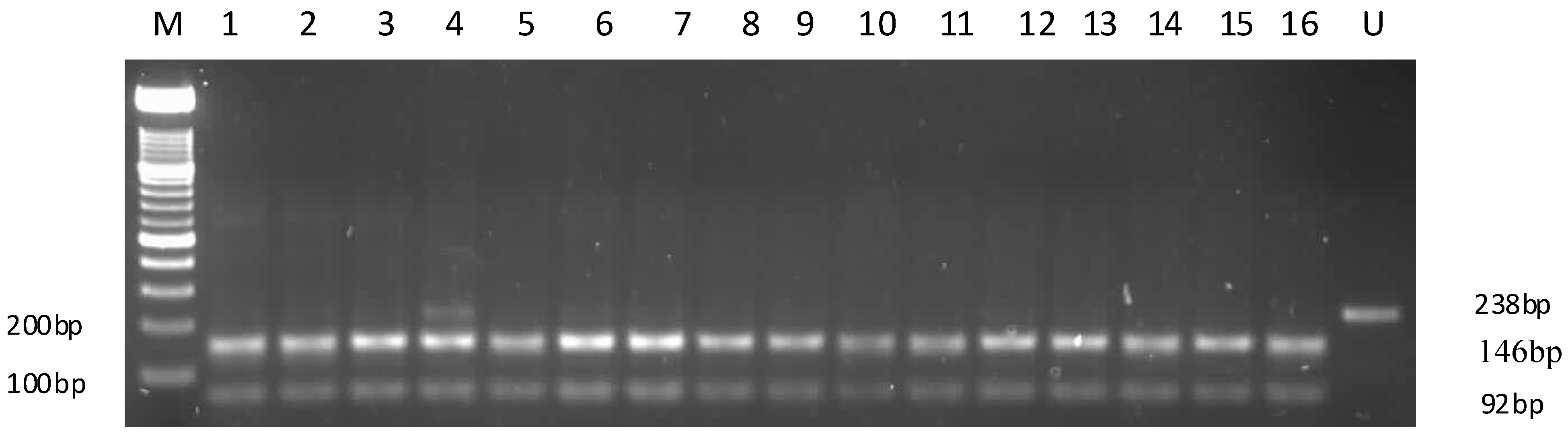
2.3. Sequencing
2.4. Genotype Frequencies
2.5. Statistical Analyses
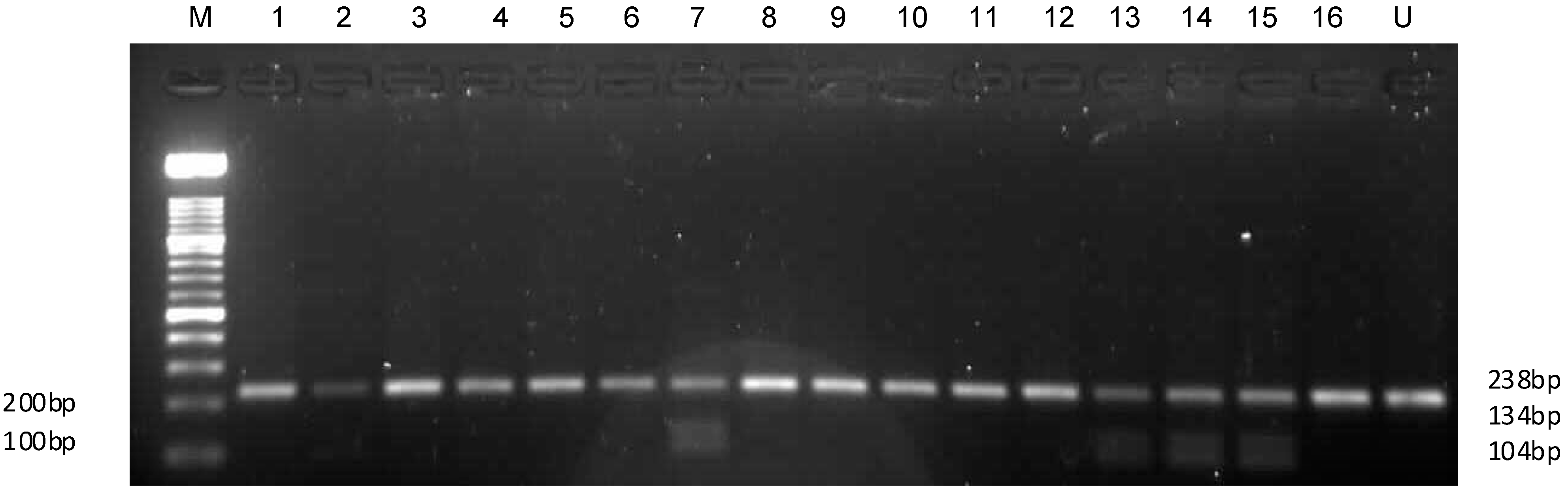
2.6. HLA Amplification and Typing Results
| APOC3 Genotypes | Mauchly’s Test for Sphericity | Repeated Measures | Mauchly’s Test for Sphericity | Repeated Measures | ||||
|---|---|---|---|---|---|---|---|---|
| CD4 | CD4 | VLD | VLD | |||||
| χ2 | p Value | F Value | p Value | χ2 | p Value | p Value | F Value | |
| −482CC | 4.1177 | 0.1276 | 56.15 | <0.0001 | 217.366 | 0.0001 | <0.0001 | 21.48 |
| −482CT | NA | NA | NA | NA | NA | NA | NA | NA |
| −482TT | NA | NA | NA | NA | NA | NA | NA | NA |
| −455TT | 3.1254 | 0.2096 | 32.25 | <0.0001 | 92.1284 | <0.0001 | 0.0119 | 8.03 |
| −455CC | 4.0513 | 0.1319 | 32.68 | <0.0001 | 99.7070 | <0.0001 | <0.0001 | 25.02 |
| Genotypes | Observed Genotypes | Expected Genotypes | Genotype Frequency | χ2 | p Value | Alleles | Expected Alleles | Allele Frequency |
|---|---|---|---|---|---|---|---|---|
| APOC3 −482CC | 196 | 194.0 | 0.985 | 126.5511103 | 0.00 | APOC3 −482C | 0.99 | 0.9875 |
| APOC3 −482CT | 01 | 4.9 | 0.005 | APOC3 −482T | 0.01 | 0.0125 | ||
| APOC3 −482TT | 02 | 0.0 | 0.010 | APOC3 −455T | 0.65 | 0.65 | ||
| APOC3 −455TT | 69 | 23.9 | 0.35 | 199 | 0.00 | APOC3 −455C | 0.35 | 0.35 |
| APOC3 −455TC | 0 | 90.2 | 0 | |||||
| APOC3 −455CC | 130 | 84.9 | 0.65 |
3. Discussion
4. Experimental Section
4.1. Study Population and DNA Extraction
4.2. Amplification of the APOC3 Promoter Region
4.3. Genotyping of APOC3 Promoter Region
4.4. HLA-B57 Gene Amplification, Sequencing, and Analysis
4.5. Statistical Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tarr, P.E.; Telenti, A. Toxicogenetics of antiretroviral therapy: Genetic factors that contribute to metabolic complications. Antivir. Ther. 2007, 12, 999–1013. [Google Scholar]
- Poma, B.Z.; Riva, A.; Nasi, M.; Cicconi, P.; Brogini, V.; Lepri, A.C.; Mologni, D.; Mazzota, F.; Monorte, A.; Mussini, C.; et al. Genetic polymorphisms differently influencing the emergence of atrophy and fat accumulation in HIV-related lipodystrophy. AIDS 2008, 22, 1769–1778. [Google Scholar] [CrossRef]
- Martin, A.; Emery, S. Metabolic disorders and cardiovascular consequences of HIV infection and antiretroviral infection. Expert Rev. Clin. Pharmacol. 2009, 2, 381–390. [Google Scholar] [CrossRef]
- Marx, B.; Sherer, R. Management of the adverse effects of antiretroviral therapy and medication adherence. Clin. Infect. Dis. 2000, 30, S96–S116. [Google Scholar] [CrossRef]
- Pirmohamed, M. Genetic factors in the predisposition to drug-induced hypersensitivity reactions. APPS J. 2006, 8, E20–E26. [Google Scholar]
- Mahungu, T.W.; Johnson, M.A.; Owen, A.; Back, D.J. The impact of pharmacogenetics on HIV therapy. Int. J. STD AIDS 2009, 20, 145–151. [Google Scholar] [CrossRef]
- Tozzi, V. Pharmacogenetics of antiretrovirals. Antivir. Res. 2010, 85, 190–200. [Google Scholar]
- Guettier, J.M.; Georgopoulos, A.; Tsai, M.Y.; Radha, V.; Shanthirani, S.; Deepa, R.; Gross, M.; Rao, G.; Mohan, V. Polymorphisms in the fatty acid-binding protein 2 and apolipoprotein C-III genes are associated with the metabolic syndrome and dyslipidemia in a South Indian population. J. Clin. Endocrinol. Metab. 2005, 90, 1705–1711. [Google Scholar]
- Clarke, H.; Mousa, S.A. The implications of pharmacogenomics in the treatment of HIV-1 infected patients of African descents. Pharmacogenomics Pers. Med. 2009, 2, 93–99. [Google Scholar]
- Tarr, P.E.; Taffe, P.; Bleiber, G.; Furrer, H.; Rotger, M.; Martinez, R.; Hirschel, B.; Battegay, M.; Weber, R.; Vernazza, P.; et al. Modeling the influence of APOC3, APOE, and TNF polymorphisms on the risk of antiretroviral therapy-associated lipid disorders. J. Infect. Dis. 2005, 191, 1419–1426. [Google Scholar] [CrossRef]
- Groenendijk, M.; Cantor, R.M.; Blom, N.H.; Rotter, J.I.; de Bruin, T.W.; Dallinga-Thie, G.M. Association of plasma lipids and apolipoproteins with the insulin response element in the apoC-III promoter region in familial combined hyperlipidemia. J. Lipid Res. 1999, 40, 1036–1044. [Google Scholar]
- Green, M.L. Evaluation and management of dyslipidemia in patients with HIV infection. J. Gen. Intern. Med. 2002, 17, 707–810. [Google Scholar] [CrossRef]
- Bonnet, E.; Ruidavets, J.; Tuech, J.; Ferrieres, J.; Collet, X.; Fauvel, J.; Massip, P.; Perret, B. Apolipoprotein C-III and E-containing lipoparticles are markedly increased in HIV-infected patients treated with protease inhibitors: Association with the development of lipodystrophy. J. Clin. Endocrinol. Metab. 2005, 86, 296–302. [Google Scholar]
- Tarr, P.E.; Rotger, M.; Telenti, A. Pharmacogenetics of dyslipidemia in HIV: Candidate gene studies of dyslipidemia in HIV-infected individuals. Pharmacogenomics 2010, 11, 587–594. [Google Scholar] [CrossRef]
- Boettiger, D. The Clinical Utility of Pharmacogenetic Tests is HIV Therapy. Available online: http://www.google.ca/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&ved=0CCsQFjAA&url=http%3a%2f%2fdspace.library.uu.nl%2fbitstream%2fhandle%2f1874%2f203507%2fPharmacogenetics_of_HIV_Antiretrovirals_-_Edited_VI%255B1%255D.doc%3fsequence%3d1&ei=CJ5LU_f7EsfmywOe14LYDg&usg=AFQjCNEUkyLNx5mNXEOLOVnAU4BAvNmNVA&sig2=IuGTzlA4IdVWyjoEgH07pQ&bvm=bv.64542518,d.bGE&cad=rjt (accessed on 14 April 2014).
- Telenti, A. Pharmacogenomics in HIV disease. In Pharmacogenomics and Personalized Medicine; Cohen, N., Ed.; Humana Press: New York, NY, USA, 2008; pp. 395–412. [Google Scholar]
- Lucas, A.; Nolan, D.; Mallal, S. HLA-B*5701 screening for susceptibility to abacavir hypersensitivity. J. Antimicrob. Chemother. 2007, 59, 591–593. [Google Scholar] [CrossRef]
- Rodriguez-Novoa, S.; Garcia-Gasco, P.; Blanco, F.; Gonzalez-Pardo, G.; Castellares, C.; Moreno, V.; Jimenez-Nacher, I.; Gonzalez-Lahoz, J.; Soriano, V. Value of the HLA-B*5701 allele to predict abacavir hypersensitivity in Spaniards. AIDS Res. Hum. Retroviruses 2007, 23, 1374–1376. [Google Scholar] [CrossRef]
- Mallal, S.; Phillips, E.; Carosi, G.; Molina, J.; Workman, C.; Tomazic, J.; Jagel-Guedes, E.; Rugina, S.; Kozyrev, O.; Cid, J.F.; et al. HLA-B*5701 screening for hypersensitivity to abacavir. N. Engl. J. Med. 2008, 358, 568–579. [Google Scholar] [CrossRef]
- Foulkes, A.S.; Wohl, D.A.; Frank, I.; Puleo, E.; Restine, S.; Wolfe, M.L.; Dube, M.P.; Tebas, P.; Reilly, M.P. Associations among race/ethnicity, APOC-III genotypes, and lipids in HIV-1 infected individuals on antiretroviral therapy. PLoS Med. 2006, 3, 337–347. [Google Scholar] [CrossRef]
- Fauvel, J.B.; Bonnet, E.; Ruidavets, J.; Ferrieres, J.; Toffoletti, A.; Massip, P.; Periet, B. An interaction between APOC-III variants and protease inhibitors contributes to high triglyceride/low HDL level in treated HIV patients. AIDS 2001, 15, 2397–2406. [Google Scholar] [CrossRef]
- Waterworth, D.M.; Talmud, P.J.; Humphries, S.E.; Wicks, P.D.; Sagnella, G.A.; Strazzullo, P.; Alberti, K.G.; Cook, D.G.; Cappuccio, F.P. Variable effects of the APOC3 –482C>T variant on insulin, glucose and triglyceride concentrations in different ethnic groups. Diabetologia 2001, 44, 245–248. [Google Scholar]
- Miller, M.; Rhyne, J.; Chen, H.; Beach, V.; Ericson, R.; Luthra, K.; Dwivedi, M.; Misra, A. APOC3 promoter polymorphisms C-482T and T-455C are associated with metabolic syndrome. Arch. Med. Res. 2007, 38, 444–451. [Google Scholar]
- Hegele, R.A.; Ban, M.R.; Carrington, C.V.; Ramdath, D.; Dan, D. Allele frequencies for candidate genes in atherosclerosis and diabetes among Trinidadian neonates. Hum. Biol. 2001, 73, 525–531. [Google Scholar] [CrossRef]
- Olivieri, O.; Martinelli, N.; Sandri, M.; Bassi, A.; Guarini, P.; Trabetti, E.; Pissolo, F.; Girelli, D.; Friso, S.; Pignatti, P.F.; et al. Apolipoprotein C-III n-3 polyunsaturated fatty acids, and “insulin-resistant” T-455 APOC3 gene polymorphism in heart disease patients: Example of gene-diet interaction. Clin. Chem. 2005, 51, 360–367. [Google Scholar] [CrossRef]
- Petersen, K.F.; Dufour, S.; Hariri, A.; Nelson-Williams, C.; Foo, J.N.; Zhang, X.; Dziura, J.; Lifton, R.P.; Shulman, G.I. Apolipoprotein C3 gene variants in nonalcoholoc fatty liver disease. N. Engl. J. Med. 2010, 362, 1082–1089. [Google Scholar] [CrossRef]
- Yu, J.; Wang, H.; Yang, S.; Yuan, J.; Chen, L.; Chen, C.L.; Huang, D.F.; Wang, Y.; Ju, S.Q.; Zhu, J. The Effect of APOC3 promoter polymorphisms on the risk of hypertriglyceridemia in chinese han population with or without type 2 diabetes mellitus. Lab Med. 2010, 41, 34–39. [Google Scholar] [CrossRef]
- Williams, F.; Meenagh, A.; Darke, C.; Acosta, A.; Daar, A.S.; Gorodezky, C.; Hammond, E.; Nascimento, E.; Middleton, S. Analysis of the distribution of HLA-B alleles in populations from five continents. Hum. Immunol. 2001, 62, 645–650. [Google Scholar] [CrossRef]
- Luo, M.; Embree, J.; Ramdahin, S.; Ndinya-Achola, J.; Njenga, S.; Bwayo, J.B.; Pan, S.; Mao, X.; Cheang, M.; Stuart, T.; et al. HLA-A and HLA-B in Kenya, Africa: Allele frequencies and identification of HLA-B*1567 and HLA-B*4426. Tissue Antigens 2002, 59, 370–380. [Google Scholar] [CrossRef]
- Paximadis, M.; Mathebula, T.Y.; Gentle, N.L.; Vardas, E.; Colvin, M.; Gray, C.M.; Tiemessen, C.T.; Puren, A. Human leukocyte antigen class I (A, B, C) and II (DRB1) diversity in the black and Caucasian South African population. Hum. Immunol. 2012, 73, 80–92. [Google Scholar]
- Ellis, J.M.; Mack, S.J.; Leke, R.F.; Quakyi, I.; Johnson, A.H.; Hurley, C.K. Diversity is demonstrated in class I HLA-A and HLA-B alleles in Cameroon, Africa: Description of HLA-A*03012, *2612, *3006 and HLA-B*1403, *4016, *4703. Tissue Antigens 2000, 56, 291–302. [Google Scholar]
- Tian, W.; Boggs, D.A.; Uko, G.; Essiet, A.; Inyama, M.; Banjoko, B.; Adewole, T.; Ding, W.Z.; Mohseni, M.; Fritz, R.; et al. MICA, HLA-B haplotypic variation in five population groups of sub-Saharan African ancestry. Genes Immun. 2003, 4, 500–505. [Google Scholar]
- Adams, S.D.; Barracchini, K.C.; Chen, D.; Robbins, F.; Wang, L.; Larsen, P.; Luhm, R.; Stroncek, D.F. Ambiguous allele combinations in HLA Class I and Class II sequence-based typing: When precise nucleotide sequencing leads to imprecise allele identification. J. Transl. Med. 2004, 2, 30. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.A.; Vilar, F.J.; Alfirevic, A.; Pirmohamed, M. Cost-effectiveness analysis of HLA B*5701 genotyping in preventing abacavir hypersensitivity. Pharmacogenetics 2004, 14, 335–342. [Google Scholar] [CrossRef]
- Zucman, D.; Truchis, P.; Majerholc, C.; Stegman, S.; Caillat-Zucman, S. Prospective screening for human leukocyte antigen-B*5701 avoids abacavir hypersensitivity reaction in the ethnically mixed French HIV population. J. Acquir. Immune Defic. Syndr. 2007, 45, 1–3. [Google Scholar] [CrossRef]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2007, 466, 707–713. [Google Scholar]
- Masebe, T.M.; Bessong, P.O.; Nwobegahay, J.; Ndip, R.N.; Meyer, D. Prevalence of MDR1 C3435T and CYP2B6 G516T polymorphisms among HIV-1 infected South African patients. Dis. Markers 2012, 32, 43–50. [Google Scholar] [CrossRef]
- BioEdit Programme. Available online: http://www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed on 14 April 2014).
- Hardy-Weinberg Equilibrium. Available online: http://www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls (accessed on 14 April 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Masebe, T.; Bessong, P.O.; Ndip, R.N.; Meyer, D. Genetic Variants of APOC3 Promoter and HLA-B Genes in an HIV Infected Cohort in Northern South Africa: A Pilot Study. Int. J. Mol. Sci. 2014, 15, 11403-11415. https://doi.org/10.3390/ijms150711403
Masebe T, Bessong PO, Ndip RN, Meyer D. Genetic Variants of APOC3 Promoter and HLA-B Genes in an HIV Infected Cohort in Northern South Africa: A Pilot Study. International Journal of Molecular Sciences. 2014; 15(7):11403-11415. https://doi.org/10.3390/ijms150711403
Chicago/Turabian StyleMasebe, Tracy, Pascal Obong Bessong, Roland Ndip Ndip, and Debra Meyer. 2014. "Genetic Variants of APOC3 Promoter and HLA-B Genes in an HIV Infected Cohort in Northern South Africa: A Pilot Study" International Journal of Molecular Sciences 15, no. 7: 11403-11415. https://doi.org/10.3390/ijms150711403




