Functional Polymorphisms of the ABCG2 Gene Are Associated with Gout Disease in the Chinese Han Male Population
Abstract
:Background
Methods
Results
Conclusion
1. Introduction
2. Results
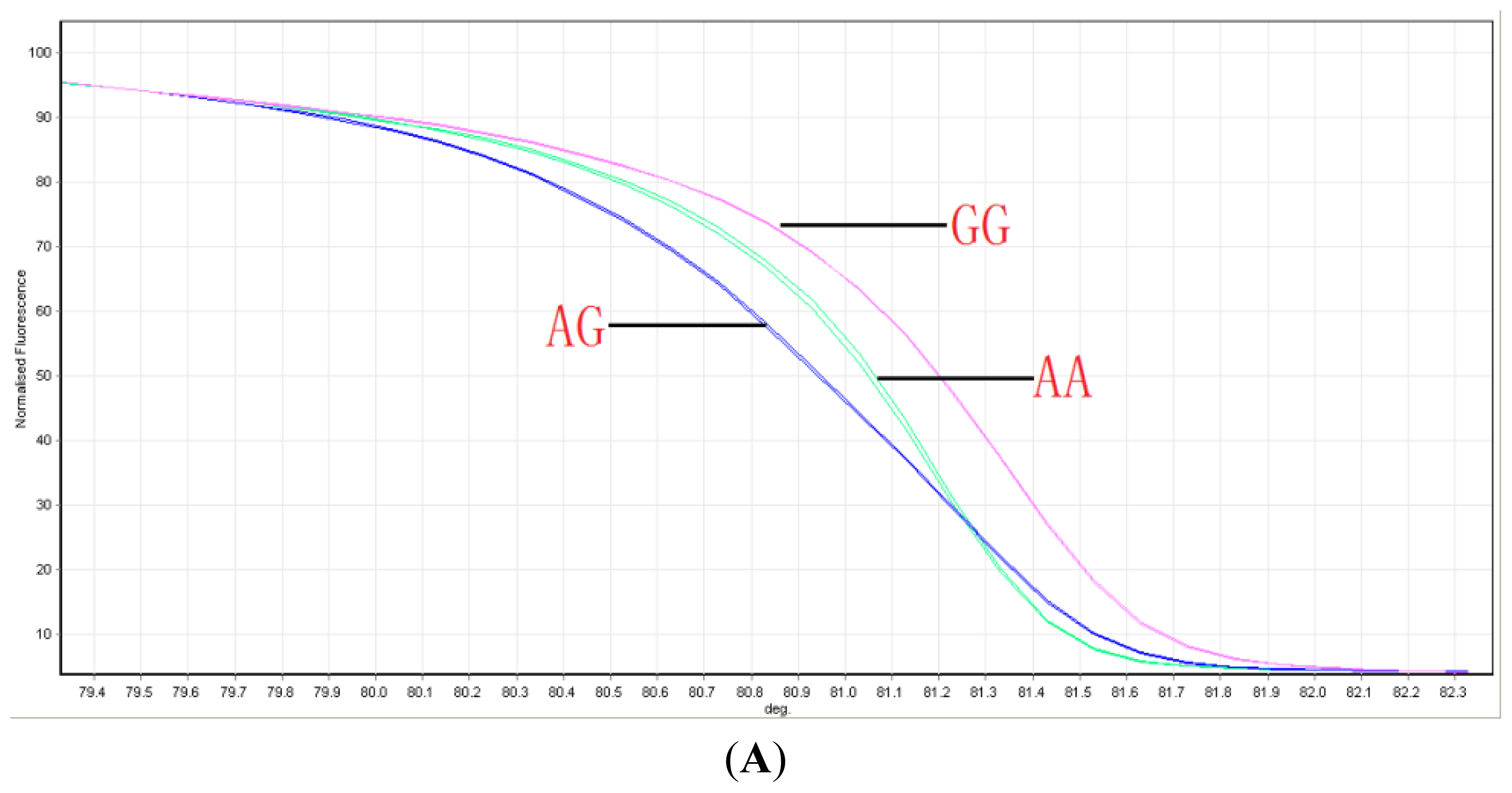
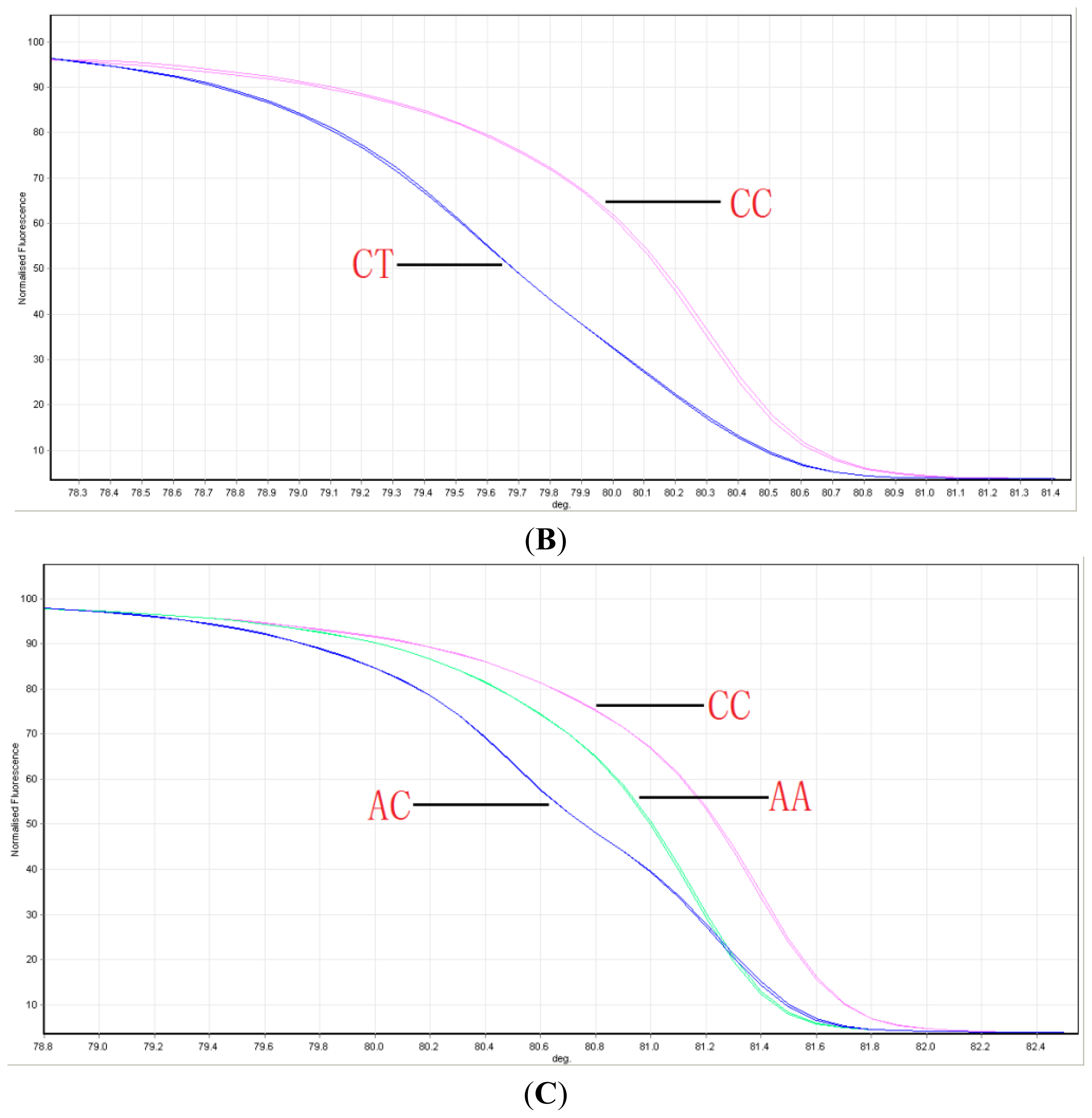
2.1. Distribution of ABCG2 Genotypes
2.2. Haplotype Analysis
2.3. Association Analysis of ABCG2 Genotype Combinations in Gout Patients
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Genotyping ABCG2 with HRM
4.3. Sequencing
4.4. Statistical Analysis
5. Conclusions
Acknowledgments
Conflicts of Interest
- Author ContributionsHejian Zou and Ming Guan designed the experiments and wrote manuscript. Danqiu Zhou, Yunqing Liu, Xinju Zhang, Hua Wang and Xiaoye Gu did the experiments, Xinhua Luo and Jin Zhang analyzed the data.
References
- Choi, H.K.; Mount, D.B.; Reginato, A.M. Pathogenesis of gout. Ann. Intern. Med 2005, 143, 499–516. [Google Scholar]
- Zhu, Y.; Pandya, B.J.; Choi, H.K. Prevalence of gout and hyperuricemia in the US general population: The National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheumatol 2011, 63, 3136–3141. [Google Scholar]
- Krishnan, E.; Lienesch, D.; Kwoh, C.K. Gout in ambulatory care settings in the United States. J. Rheumatol 2008, 35, 498–501. [Google Scholar]
- Trieste, L.; Palla, I.; Fusco, F.; Tani, C.; Baldini, C.; Mosca, M.; Turchetti, G. The economic impact of gout: A systematic literature review. Clin. Exp. Rheumatol 2012, 30, S145–S148. [Google Scholar]
- Tuttle, K.R.; Short, R.A.; Johnson, R.J. Sex differences in uric acid and risk factors for coronary artery disease. Am. J. Cardiol 2001, 87, 1411–1414. [Google Scholar]
- Talaat, K.M.; el-Sheikh, A.R. The effect of mild hyperuricemia on urinary transforming growth factor beta and the progression of chronic kidney disease. Am. J. Nephrol 2007, 27, 435–440. [Google Scholar]
- Nath, S.D.; Voruganti, V.S.; Arar, N.H.; Thameem, F.; Lopez-Alvarenga, J.C.; Bauer, R.; Blangero, J.; MacCluer, J.W.; Comuzzie, A.G.; Abboud, H.E. Genome scan for determinants of serum uric acid variability. J. Am. Soc. Nephrol 2007, 18, 3156–3163. [Google Scholar]
- Enomoto, A.; Kimura, H.; Chairoungdua, A.; Shigeta, Y.; Jutabha, P.; Cha, S.H.; Hosoyamada, M.; Takeda, M.; Sekine, T.; Igarashi, T.; et al. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature 2002, 417, 447–452. [Google Scholar]
- Vitart, V.; Rudan, I.; Hayward, C.; Gray, N.K.; Floyd, J.; Palmer, C.N.; Knott, S.A.; Kolcic, I.; Polasek, O.; Graessler, J.; et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat. Genet 2008, 40, 437–442. [Google Scholar]
- Kolz, M.; Johnson, T.; Sanna, S.; Teumer, A.; Vitart, V.; Perola, M.; Mangino, M.; Albrecht, E.; Wallace, C.; Farrall, M.; et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet 2009, 5, e1000504. [Google Scholar]
- Cheng, L.S.; Chiang, S.L.; Tu, H.P.; Chang, S.J.; Wang, T.N.; Ko, A.M.; Chakraborty, R.; Ko, Y.C. Genomewide scan for gout in taiwanese aborigines reveals linkage to chromosome 4q25. Am. J. Hum. Genet 2004, 75, 498–503. [Google Scholar]
- Woodward, O.M.; Kottgen, A.; Coresh, J.; Boerwinkle, E.; Guggino, W.B.; Kottgen, M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc. Natl. Acad. Sci. USA 2009, 106, 10338–10342. [Google Scholar]
- Ichida, K.; Matsuo, H.; Takada, T.; Nakayama, A.; Murakami, K.; Shimizu, T.; Yamanashi, Y.; Kasuga, H.; Nakashima, H.; Nakamura, T.; et al. Decreased extra-renal urate excretion is a common cause of hyperuricemia. Nat. Commun 2012, 3, 764. [Google Scholar]
- Tamura, A.; Wakabayashi, K.; Onishi, Y.; Takeda, M.; Ikegami, Y.; Sawada, S.; Tsuji, M.; Matsuda, Y.; Ishikawa, T. Re-evaluation and functional classification of non-synonymous single nucleotide polymorphisms of the human ATP-binding cassette transporter ABCG2. Cancer Sci 2007, 98, 231–239. [Google Scholar]
- Dehghan, A.; Kottgen, A.; Yang, Q.; Hwang, S.J.; Kao, W.L.; Rivadeneira, F.; Boerwinkle, E.; Levy, D.; Hofman, A.; Astor, B.C.; et al. Association of three genetic loci with uric acid concentration and risk of gout: A genome-wide association study. Lancet 2008, 372, 1953–1961. [Google Scholar]
- Matsuo, H.; Ichida, K.; Takada, T.; Nakayama, A.; Nakashima, H.; Nakamura, T.; Kawamura, Y.; Takada, Y.; Yamamoto, K.; Inoue, H.; et al. Common dysfunctional variants in ABCG2 are a major cause of early-onset gout. Sci. Rep 3.
- Mizuarai, S.; Aozasa, N.; Kotani, H. Single nucleotide polymorphisms result in impaired membrane localization and reduced atpase activity in multidrug transporter ABCG2. Int. J. Cancer 2004, 109, 238–246. [Google Scholar]
- Matsuo, H.; Takada, T.; Ichida, K.; Nakamura, T.; Nakayama, A.; Ikebuchi, Y.; Ito, K.; Kusanagi, Y.; Chiba, T.; Tadokoro, S.; et al. Common defects of ABCG2, a high-capacity urate exporter, cause gout: A function-based genetic analysis in a Japanese population. Sci. Transl. Med 2009, 1, 5ra11. [Google Scholar]
- Ishikawa, T.; Aw, W.; Kaneko, K. Metabolic Interactions of purine derivatives with human ABC transporter ABCG2: Genetic testing to assess gout risk. Pharmaceuticals (Basel) 2013, 6, 1347–1360. [Google Scholar]
- Endou, H.; Anzai, N. Urate transport across the apical membrane of renal proximal tubules. Nucleosides Nucleotides Nucleic Acids 2008, 27, 578–584. [Google Scholar]
- Anzai, N.; Jutabha, P.; Amonpatumrat-Takahashi, S.; Sakurai, H. Recent advances in renal urate transport: Characterization of candidate transporters indicated by genome-wide association studies. Clin. Exp. Nephrol 2012, 16, 89–95. [Google Scholar]
- George, R.L.; Keenan, R.T. Genetics of hyperuricemia and gout: Implications for the present and future. Curr. Rheumatol. Rep 2013, 15, 309. [Google Scholar]
- Guan, M.; Zhou, D.; Ma, W.; Chen, Y.; Zhang, J.; Zou, H. Association of an intronic SNP of SLC2A9 gene with serum uric acid levels in the Chinese male Han population by high-resolution melting method. Clin. Rheumatol 2011, 30, 29–35. [Google Scholar]
- Li, C.; Chu, N.; Wang, B.; Wang, J.; Luan, J.; Han, L.; Meng, D.; Wang, Y.; Suo, P.; Cheng, L.; et al. Polymorphisms in the presumptive promoter region of the SLC2A9 gene are associated with gout in a Chinese male population. PLoS One 2012, 7, e24561. [Google Scholar]
- Li, C.; Han, L.; Levin, A.M.; Song, H.; Yan, S.; Wang, Y.; Meng, D.; Lv, S.; Ji, Y.; Xu, X.; et al. Multiple single nucleotide polymorphisms in the human urate transporter 1 (hURAT1) gene are associated with hyperuricaemia in Han Chinese. J. Med. Genet 2010, 47, 204–210. [Google Scholar]
- Guan, M.; Zhang, J.; Chen, Y.; Liu, W.; Kong, N.; Zou, H. High-resolution melting analysis for the rapid detection of an intronic single nucleotide polymorphism in SLC22A12 in male patients with primary gout in China. Scand. J. Rheumatol 2009, 38, 276–281. [Google Scholar]
- Hosomi, A.; Nakanishi, T.; Fujita, T.; Tamai, I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PLoS One 2012, 7, e30456. [Google Scholar]
- Woodward, O.M.; Tukaye, D.N.; Cui, J.; Greenwell, P.; Constantoulakis, L.M.; Parker, B.S.; Rao, A.; Kottgen, M.; Maloney, P.C.; Guggino, W.B. Gout-causing Q141K mutation in ABCG2 leads to instability of the nucleotide-binding domain and can be corrected with small molecules. Proc. Natl. Acad. Sci. USA 2013, 110, 5223–5228. [Google Scholar]
- Zhang, L.; Spencer, K.L.; Voruganti, V.S.; Jorgensen, N.W.; Fornage, M.; Best, L.G.; Brown-Gentry, K.D.; Cole, S.A.; Crawford, D.C.; Deelman, E.; et al. Association of functional polymorphism rs2231142 (Q141K) in the ABCG2 gene with serum uric acid and gout in 4 US populations: The PAGE study. Am. J. Epidemiol 2013, 177, 923–932. [Google Scholar]
- Lepper, E.R.; Nooter, K.; Verweij, J.; Acharya, M.R.; Figg, W.D.; Sparreboom, A. Mechanisms of resistance to anticancer drugs: The role of the polymorphic ABC transporters ABCB1 and ABCG2. Pharmacogenomics 2005, 6, 115–138. [Google Scholar]
- Wang, B.; Miao, Z.; Liu, S.; Wang, J.; Zhou, S.; Han, L.; Meng, D.; Wang, Y.; Li, C.; Ma, X. Genetic analysis of ABCG2 gene C421A polymorphism with gout disease in Chinese Han male population. Hum. Genet 2010, 127, 245–246. [Google Scholar]
- Kim, H.S.; Sunwoo, Y.E.; Ryu, J.Y.; Kang, H.J.; Jung, H.E.; Song, I.S.; Kim, E.Y.; Shim, J.C.; Shon, J.H.; Shin, J.G. The effect of ABCG2 V12M, Q141K and Q126X, known functional variants in vitro, on the disposition of lamivudine. Br. J. Clin. Pharmacol 2007, 64, 645–654. [Google Scholar]
- Itoda, M.; Saito, Y.; Shirao, K.; Minami, H.; Ohtsu, A.; Yoshida, T.; Saijo, N.; Suzuki, H.; Sugiyama, Y.; Ozawa, S.; et al. Eight novel single nucleotide polymorphisms in ABCG2/BCRP in Japanese cancer patients administered irinotacan. Drug Metab. Pharmacokinet 2005, 18, 212–217. [Google Scholar]
- Kobayashi, D.; Ieiri, I.; Hirota, T.; Takane, H.; Maegawa, S.; Kigawa, J.; Suzuki, H.; Nanba, E.; Oshimura, M.; Terakawa, N.; et al. Functional assessment of ABCG2 (BCRP) gene polymorphisms to protein expression in human placenta. Drug Metab. Dispos 2005, 33, 94–101. [Google Scholar]
- Loo, T.W.; Bartlett, M.C.; Clarke, D.M. Rescue of folding defects in ABC transporters using pharmacological chaperones. J. Bioenerg. Biomembr 2005, 37, 501–507. [Google Scholar]
- Chiba, P.; Freissmuth, M.; Stockner, T. Defining the blanks-Pharmacochaperoning of SLC6 transporters and ABC transporters. Pharmacol. Res 2013. [Google Scholar] [CrossRef]
| SNP | Genotype * | Allele Frequency Mode | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | p-Value | p-Value | OR | 95% CI | |||||||
| 1/1 | 1/2 | 2/2 | MAF | 1/1 | 1/2 | 2/2 | MAF | |||||
| Q141K | 84 | 181 | 87 | 0.496 | 33 | 150 | 167 | 0.309 | 1.18 × 10−11 | 8.99 × 10−13 | 2.20 | 1.77–2.74 |
| Q126X | 0 | 33 | 319 | 0.047 | 0 | 12 | 338 | 0.017 | 1.31 × 10−3 | 1.57 × 10−3 | 2.91 | 1.49–5.68 |
| V12M | 16 | 97 | 239 | 0.183 | 35 | 133 | 182 | 0.290 | 3.67 × 10−5 | 2.55 × 10−6 | 0.55 | 0.43–0.71 |
| Allele | Frequency | p-Value | OR | 95% CI | |||
|---|---|---|---|---|---|---|---|
| V12M | Q126X | Q141K | Gout | Control | |||
| G | C | A | 0.481 | 0.289 | 1.26 × 10−13 | 2.30 | 1.84–2.87 |
| G | T | C | 0.044 | 0.017 | 2.97 × 10−3 | 2.71 | 1.37–5.36 |
| G | C | C | 0.292 | 0.404 | 8.27 × 10−6 | 0.60 | 0.48–0.75 |
| A | C | C | 0.165 | 0.271 | 1.53 × 10−6 | 0.53 | 0.41–0.69 |
| Estimated Function | Genotype Combination | Number (%) | p-Value | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|
| Q141K | Q126X | Gout | Control | ||||
| ≤1/4 function | C/A | T/C | 22 (6.2) | 8 (2.3) | 8.47 × 10−6 | 5.90 | 2.56–13.58 |
| 1/2 function | C/C | T/C | 95 (27.0) | 37 (10.5) | 1.12 × 10−13 | 5.51 | 3.46–8.77 |
| A/A | C/C | ||||||
| 3/4 function | C/A | C/C | 159 (45.2) | 142 (40.6) | 1.00 × 10−6 | 2.40 | 1.69–3.42 |
| Full function | C/C | C/C | 76 (21.6) | 163 (46.6) | — | — | |
| Index | Gout Patients | Controls | p-Value |
|---|---|---|---|
| Subjects (%) | 352 (50.1%) | 350 (49.9%) | |
| Age (year) | 57.6 ± 14.0 | 56.6 ± 16.6 | NS |
| BUN (mmol/L) | 5.4 ± 1.9 | 5.5 ± 2.1 | NS |
| Creatinine (μmol/L) | 97.3 ± 15.7 | 96.1 ± 16.4 | NS |
| Uric Acid (μmol/L) | 456.4 ± 120.1 | 334.7 ± 88.7 | <0.01 |
| SNP ID | SNP Allele | Sequence (5′-3′) | Size |
|---|---|---|---|
| V12M | A/G | ATGGTATGGGCCATTCATTG ATGCCTTCAGGTCATTGGAA | 250 bp |
| Q141K | A/C | ATGTTGTGATGGGCACTCTG CCACATTACCTTGGAGTCTG | 158 bp |
| Q126X | C/T | GCTGCAAGGAAAGATCCAAG CAGCCAAAGCACTTACCCAT | 163 bp |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhou, D.; Liu, Y.; Zhang, X.; Gu, X.; Wang, H.; Luo, X.; Zhang, J.; Zou, H.; Guan, M. Functional Polymorphisms of the ABCG2 Gene Are Associated with Gout Disease in the Chinese Han Male Population. Int. J. Mol. Sci. 2014, 15, 9149-9159. https://doi.org/10.3390/ijms15059149
Zhou D, Liu Y, Zhang X, Gu X, Wang H, Luo X, Zhang J, Zou H, Guan M. Functional Polymorphisms of the ABCG2 Gene Are Associated with Gout Disease in the Chinese Han Male Population. International Journal of Molecular Sciences. 2014; 15(5):9149-9159. https://doi.org/10.3390/ijms15059149
Chicago/Turabian StyleZhou, Danqiu, Yunqing Liu, Xinju Zhang, Xiaoye Gu, Hua Wang, Xinhua Luo, Jin Zhang, Hejian Zou, and Ming Guan. 2014. "Functional Polymorphisms of the ABCG2 Gene Are Associated with Gout Disease in the Chinese Han Male Population" International Journal of Molecular Sciences 15, no. 5: 9149-9159. https://doi.org/10.3390/ijms15059149




