Inhibitory Effects of Benzaldehyde Derivatives from the Marine Fungus Eurotium sp. SF-5989 on Inflammatory Mediators via the Induction of Heme Oxygenase-1 in Lipopolysaccharide-Stimulated RAW264.7 Macrophages
Abstract
:1. Introduction
2. Results and Discussion
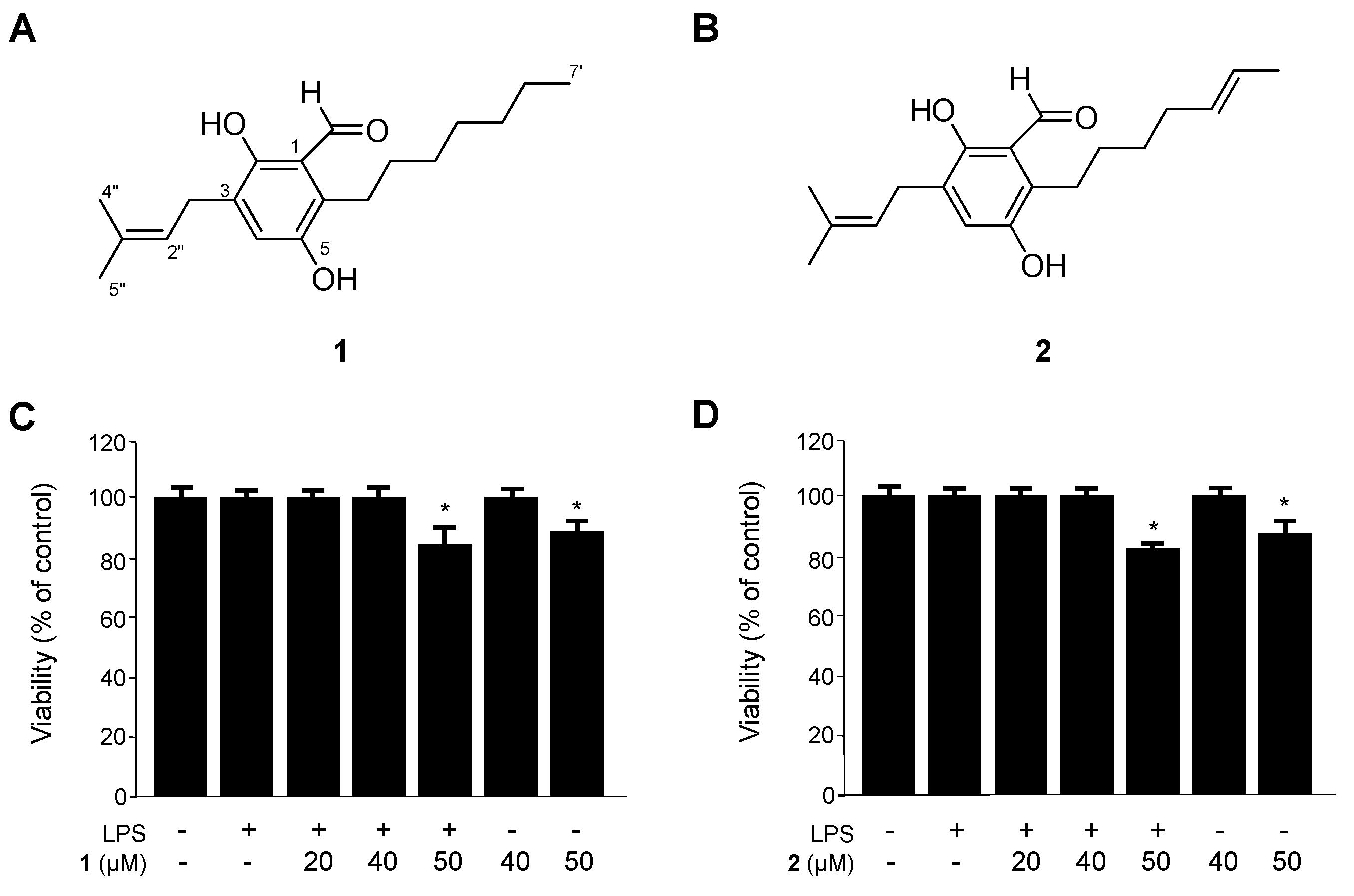
2.1. Identification of Flavoglaucin (1) and Isotetrahydro-Auroglaucin (2), and Their Effects on the Viability of RAW264.7 Macrophages
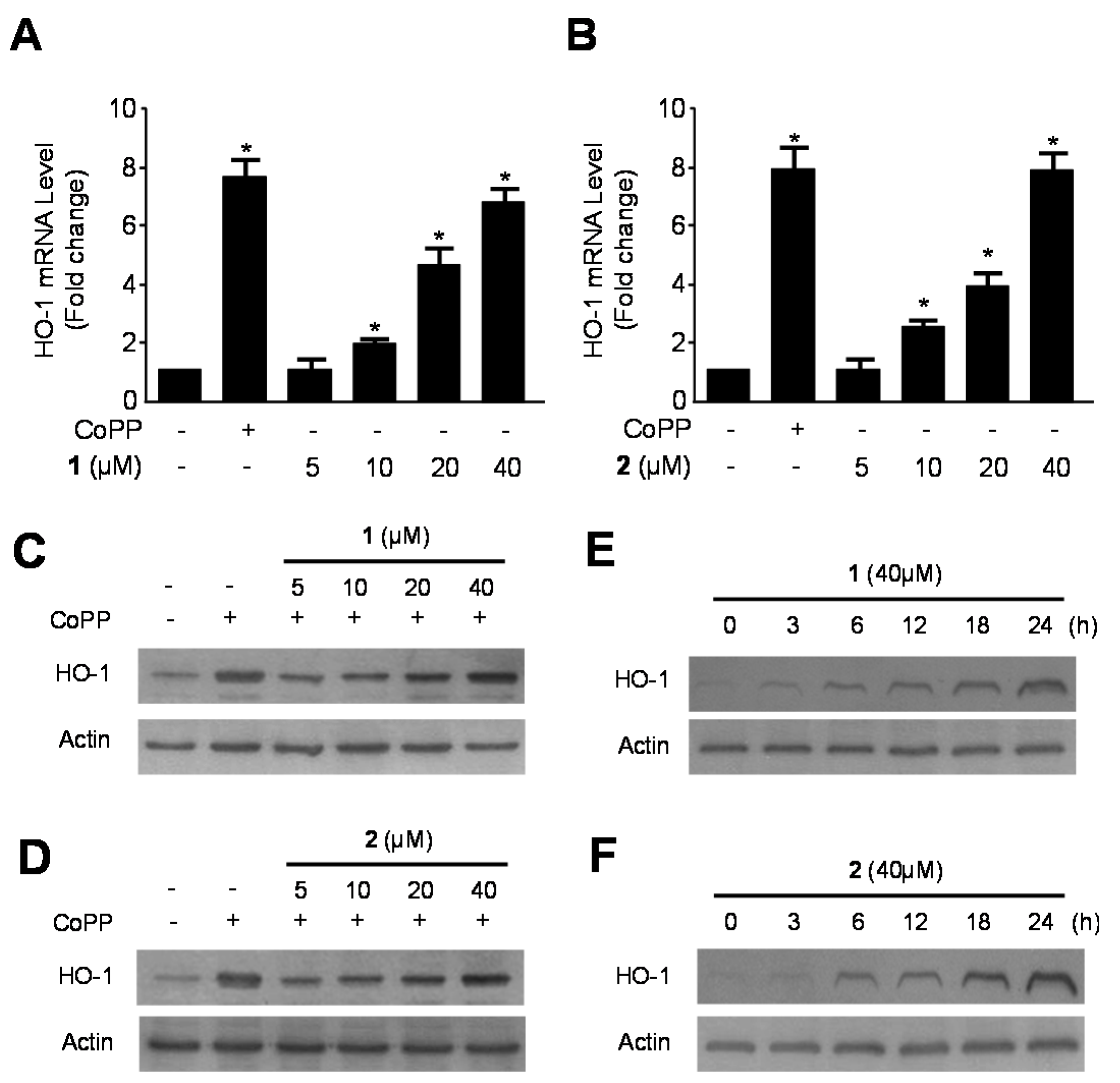
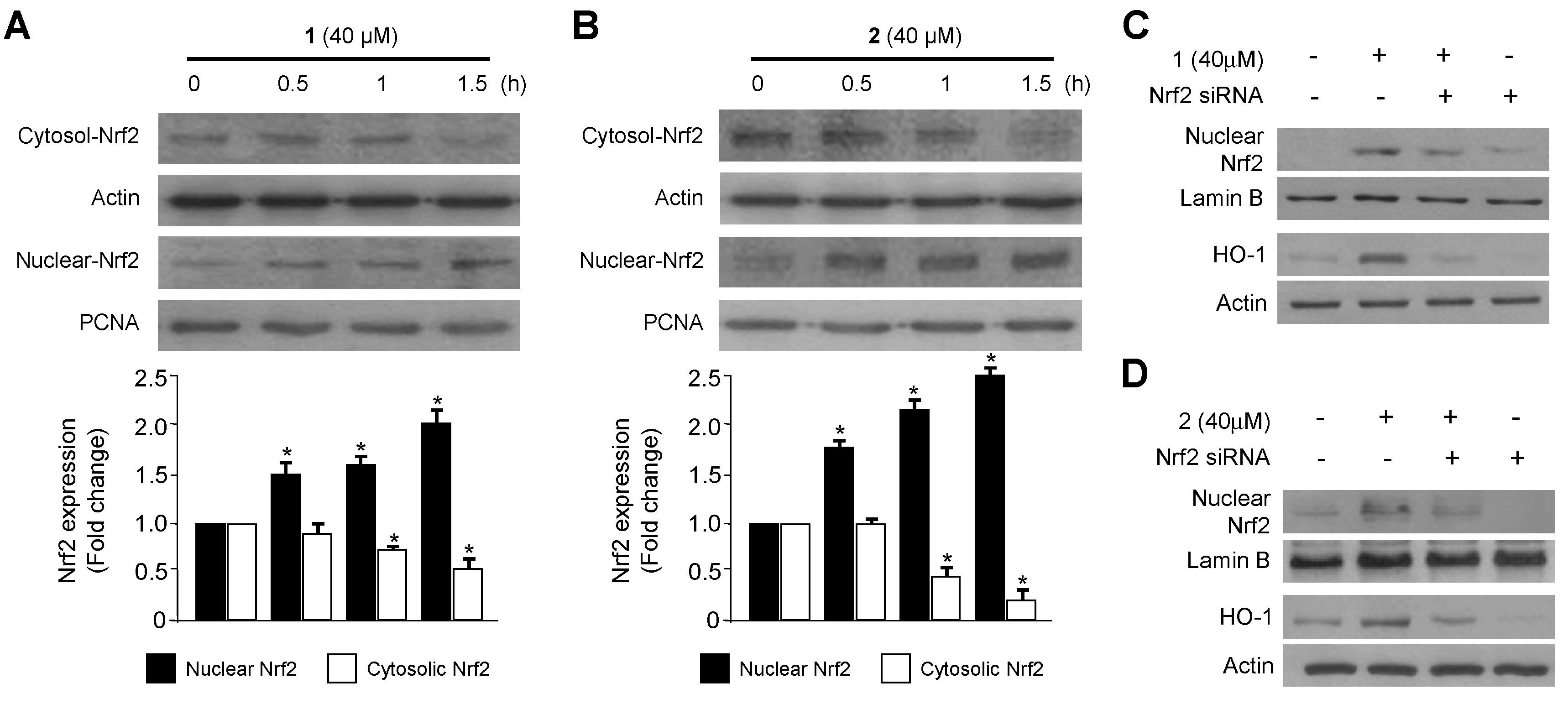
2.2. Effects of 1 and 2 on HO-1 mRNA and Protein Expression via Nuclear Translocation of Nuclear Transcription Factor-E2–Related Factor 2 (Nrf2) in RAW264.7 Macrophages
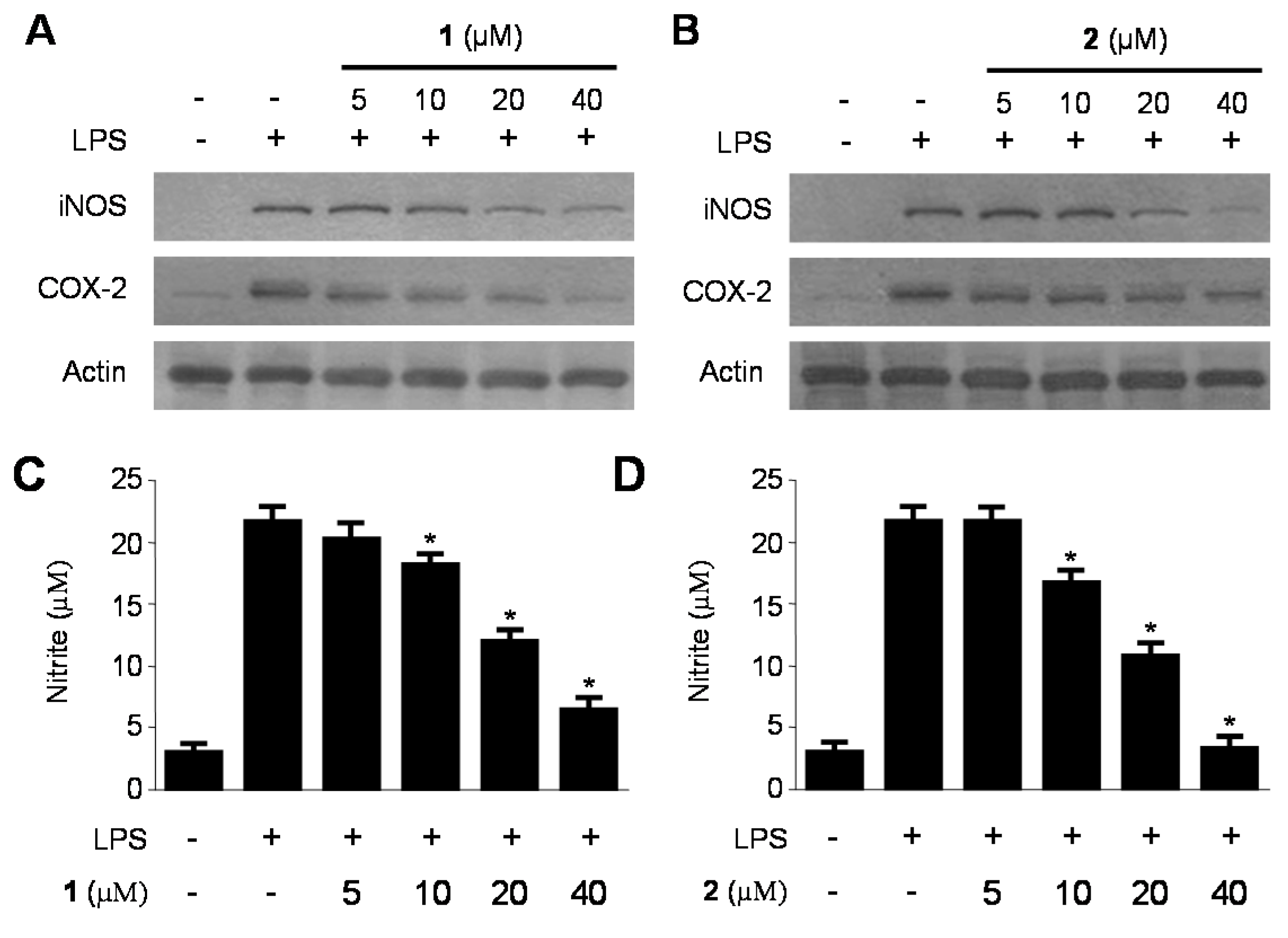
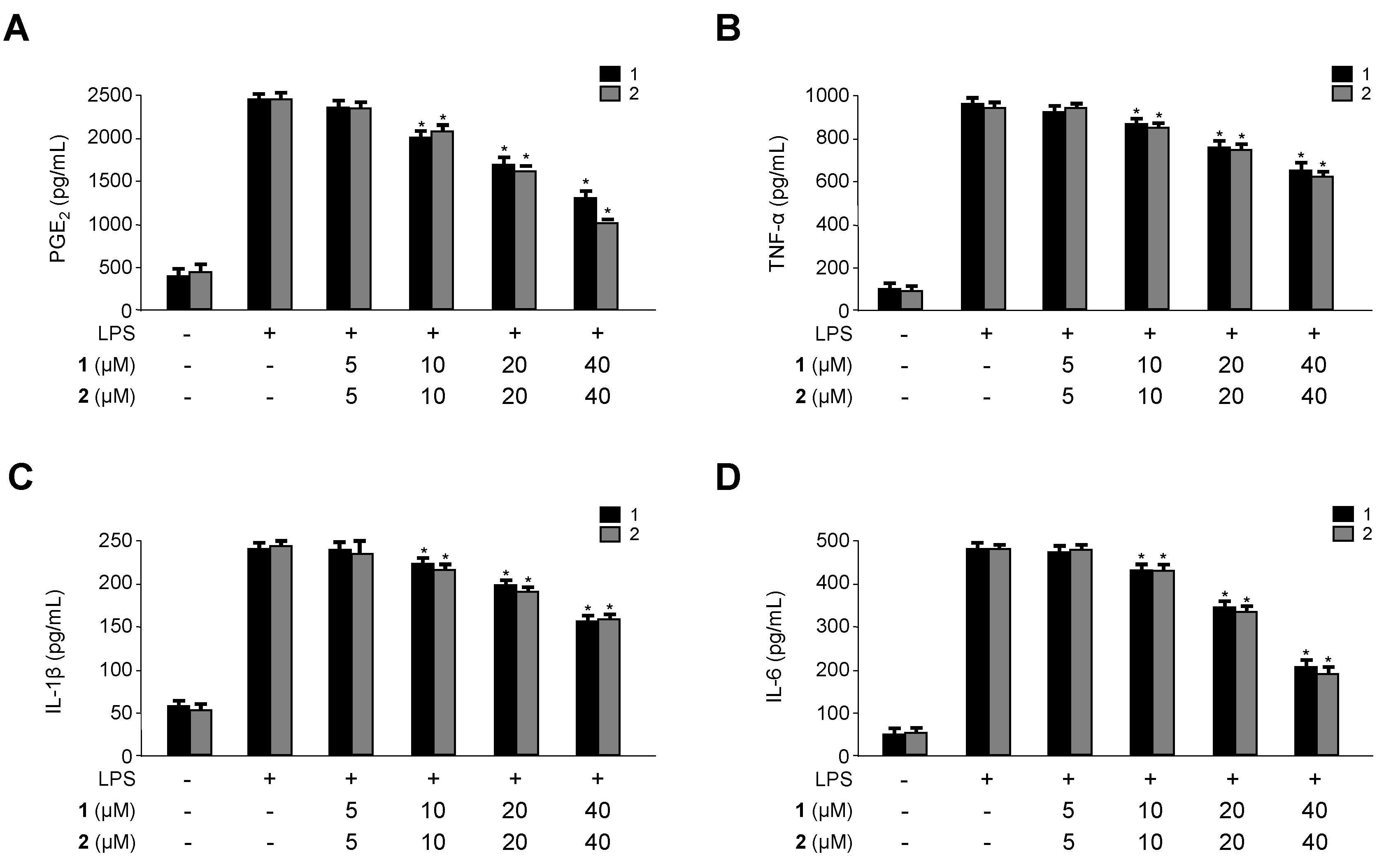
2.3. Inhibitory Effects of 1 and 2 on the Production and Expression of Pro-Inflammatory Cytokines and Mediators in LPS-Stimulated RAW264.7 Macrophages
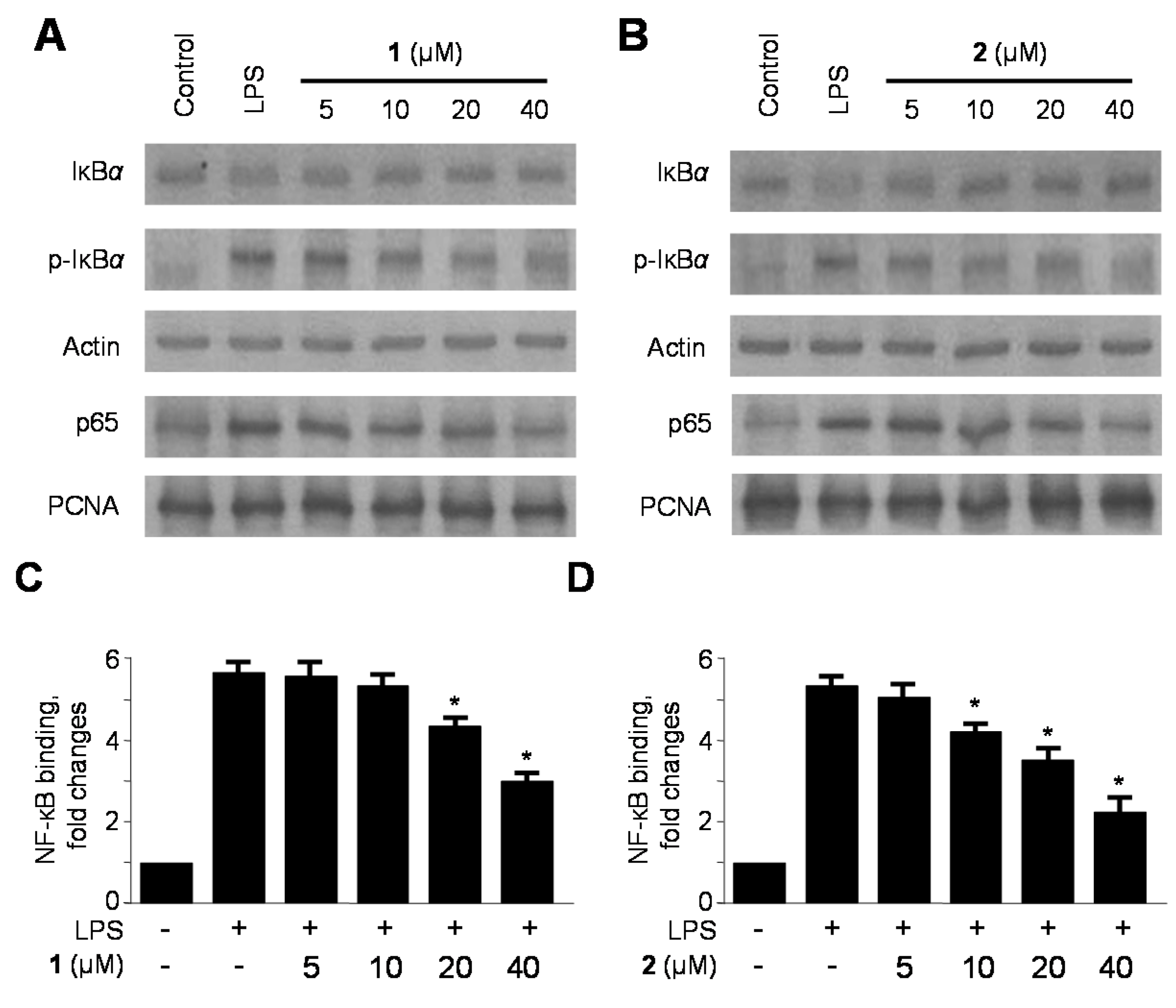
2.4. Effects of 1 and 2 on Nuclear Factor-κB (NF-κB) Activation in LPS-Stimulated RAW264.7 Macrophages
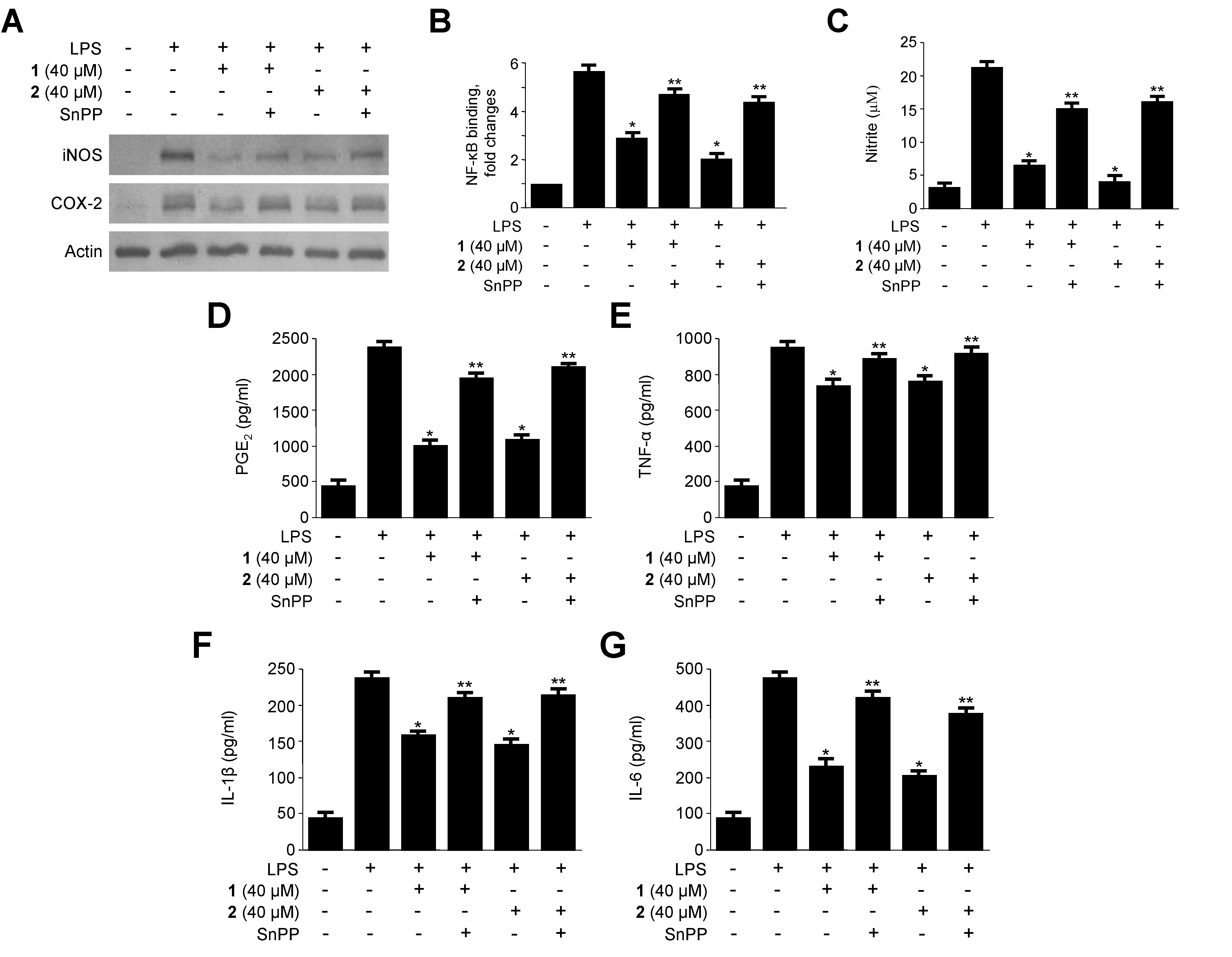
2.5. Effects of HO-1 Expression on the Inhibition of Proinflammatory Mediators, Cytokines, and NF-κB Activity by 1 and 2 in LPS-Stimulated RAW264.7 Macrophages
3. Experimental Section
3.1. General
3.2. Specimen Collection and Identification of the Marine-Derived Fungus Eurotium sp. SF-5989
3.3. Fermentation, Extraction and Isolation from Eurotium sp. SF-5989
3.3.1. Flavoglaucin (1)
3.3.2. Isotetrahydro-Auroglaucin (2)
3.4. Cell Culture and Viability Assay
3.5. Real-Time PCR
3.6. Western Blot Analysis
3.7. Determination of Nitrite Production and PGE2, TNF-α, IL-1β and IL-6 Assays
3.8. Preparation of Cytosolic and Nuclear Fractions
3.9. DNA Binding Activity of NF-κB
3.10. Transfection
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Berenbaum, F. Proinflammatory cytokines, prostaglandins, and the chondrocyte: Mechanisms of intracellular activation. Jt. Bone Spine 2000, 67, 561–564. [Google Scholar] [CrossRef]
- Karpurapu, M.; Wang, X.; Deng, J.; Park, H.; Xiao, L.; Sadikot, R.T.; Frey, R.S.; Maus, U.A.; Park, G.Y.; Scott, E.W.; et al. Functional PU.1 in macrophages has a pivotal role in NF-κB activation and neutrophilic lung inflammation during endotoxemia. Blood 2011, 118, 5255–5266. [Google Scholar]
- McCartney-Francis, N.; Allen, J.B.; Mizel, D.E.; Albina, J.E.; Xie, Q.W.; Nathan, C.F.; Wahl, S.M. Suppression of arthritis by an inhibitor of nitric oxide synthase. J. Exp. Med. 1993, 178, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Szabó, C. Alterations in nitric oxide production in various forms of circulatory shock. New Horiz. 1995, 3, 2–32. [Google Scholar] [PubMed]
- Samuelsson, B.; Morgenstern, R.; Jakobsson, P.J. Membrane prostaglandin E synthase-1: A novel therapeutic target. Pharmacol. Rev. 2007, 59, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Simmons, D.L.; Botting, R.M.; Hla, T. Cyclooxygenase isozymes: The biology of prostaglandin synthesis and inhibition. Pharmacol. Rev. 2004, 56, 387–437. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.T.; Pae, H.O.; Cha, Y.N. Role of heme oxygenase-1 in vascular disease. Curr. Pharm. Des. 2008, 14, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.S.; Tsai, H.L.; Chau, L.Y. Induction of heme oxygenase-1 expression in murine macrophages is essential for the anti-inflammatory effect of low dose 15-deoxy-δ 12,14-prostaglandin J2. J. Biol. Chem. 2003, 278, 19325–19330. [Google Scholar] [CrossRef] [PubMed]
- Otterbein, L.E.; Bach, F.H.; Alam, J.; Soares, M.; Lu, H.T.; Wysk, M.; Davis, R.J.; Flavell, R.A.; Choi, A.M. Carbon monoxide has anti-inflammatory effects involving the mitogen-activated protein kinase pathway. Nat. Med. 2000, 6, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Wiesel, P.; Foster, L.C.; Pellacani, A.; Layne, M.D.; Hsieh, C.M.; Huggins, G.S.; Strauss, P.; Yet, S.F.; Perrella, M.A. Thioredoxin facilitates the induction of heme oxygenase-1 in response to inflammatory mediators. J. Biol. Chem. 2000, 275, 24840–24846. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.M.; Kim, M.Y.; Park, M.K.; Lee, Y.S.; Kim, Y.M.; Kim, H.J.; Lee, J.H.; Chang, K.C. Higenamine reduces HMGB1 during hypoxia-induced brain injury by induction of heme oxygenase-1 through PI3K/Akt/Nrf-2 signal pathways. Apoptosis 2012, 17, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Verma, I.M. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2002, 2, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kauppinen, A.; Kaarniranta, K. Phytochemicals suppress nuclear factor-κB signaling: Impact on health span and the aging process. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of micro-organisms review. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Bhadury, P.; Mohammad, B.T.; Wright, P.C. The current status of natural products from marine fungi and their potential as anti-infective agents. J. Ind. Microbiol. Biotechnol. 2006, 33, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Cui, X.; Lee, D.S.; Sohn, J.H.; Yim, J.H.; Kim, Y.C.; Oh, H. Anti-inflammatory effect of neoechinulin A from the marine fungus Eurotium sp. SF-5989 through the suppression of NF-кB and p38 MAPK pathways in lipopolysaccharide-stimulated RAW264.7 macrophages. Molecules 2013, 18, 13245–13259. [Google Scholar]
- Hamasaki, T.; Fukunaga, M.; Kimura, Y.; Hatsuda, Y. Isolation and structures of two new metabolites from Aspergillus ruber. Agric. Biol. Chem. 1980, 44, 1685–1687. [Google Scholar] [CrossRef]
- Podojil, M.; Sedmera, P.; Vokoun, J.; Betina, V.; Baráthová, H.; Duracková, Z.; Horáková, K.; Nemec, P. Eurotium (Aspergillus) repens metabolites and their biological activity. Folia Microbiol. 1979, 23, 438–443. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Lee, U.; Kang, J.S.; Choi, H.D.; Sona, B.W. A new radical scavenging anthracene glycoside, asperflavin ribofuranoside, and polyketides from a marine isolate of the fungus microsporum. Chem. Pharm. Bull. 2006, 54, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Ito, C.; Itoigawa, M.; Osawa, T. Antioxidants produced by Eurotium herbariorum of filamentous fungi used for the manufacture of Karebushi, dried bonito (Katsuobushi). Biosci. Biotechnol. Biochem. 2009, 73, 1323–1327. [Google Scholar] [CrossRef] [PubMed]
- Umeda, M.; Yamashita, T.; Saito, M.; Sekita, S.; Takahashi, C.; Yoshihira, K.; Natori, S.; Kurata, H.; Udagawa, S.I. Chemical and cytotoxicity survey on the metabolites of toxic fungi. Jpn. J. Exp. Med. 1974, 44, 83–96. [Google Scholar] [PubMed]
- Miyake, Y.; Ito, C.; Tokuda, H.; Osawa, T.; Itoigawa, M. Evaluation of flavoglaucin, its derivatives and pyranonigrins produced by molds used in fermented foods for inhibiting tumor promotion. Biosci. Biotechnol. Biochem. 2010, 74, 1120–1122. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; León, F.; Radwan, M.M.; Dale, O.R.; Husni, A.S.; Manly, S.P.; Lupien, S.; Wang, X.; Hill, R.A.; Dugan, F.M.; et al. Benzyl derivatives with in vitro binding affinity for human opioid and cannabinoid receptors from the fungus Eurotium repens. J. Nat. Prod. 2011, 74, 1636–1639. [Google Scholar]
- Sohn, J.H.; Lee, Y.R.; Lee, D.S.; Kim, Y.C.; Oh, H. PTP1B inhibitory secondary metabolites from marine-derived fungal strains Penicillium spp. and Eurotium sp. J. Microbiol. Biotechnol. 2013, 23, 1206–1211. [Google Scholar] [CrossRef]
- Kobayashi, H.; Takeno, M.; Saito, T.; Takeda, Y.; Kirino, Y.; Noyori, K.; Hayashi, T.; Ueda, A.; Ishigatsubo, Y. Regulatory role of heme oxygenase 1 in inflammation of rheumatoid arthritis. Arthritis Rheumatol. 2006, 54, 1132–1142. [Google Scholar] [CrossRef]
- Otterbein, L.E.; Lee, P.J.; Chin, B.Y.; Petrache, I.; Camhi, S.L.; Alam, J.; Choi, A.M. Protective effects of heme oxygenase-1 in acute lung injury. Chest 1999, 116, 61S–63S. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W.; Choi, A.M. Therapeutic applications of carbon monoxide in lung disease. Curr. Opin. Pharmacol. 2006, 6, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kyung, C.Y.; Lee, K.S.; Cho, D.H.; Baek, Y.Y.; Lee, D.K.; Ha, K.S.; Choe, J.; Won, M.H.; Jeoung, D.; et al. Functional dissection of Nrf2-dependent phase II genes in vascular inflammation and endotoxic injury using Keap1 siRNA. Free Radic. Biol. Med. 2012, 53, 629–640. [Google Scholar]
- Jun, M.S.; Kim, H.S.; Kim, Y.M.; Kim, H.J.; Park, E.J.; Lee, J.H.; Lee, K.R.; Kim, Y.S.; Chang, K.C. Ethanol extract of Prunella vulgaris var. lilacina inhibits HMGB1 release by induction of heme oxygenase-1 in LPS-activated RAW264.7 cells and CLP induced septic mice. Phytother. Res. 2012, 26, 605–612. [Google Scholar]
- Lee, D.S.; Jeong, G.S.; Li, B.; Lee, S.U.; Oh, H.; Kim, Y.C. Asperlin from the marine-derived fungus Aspergillus sp. SF-5044 exerts anti-inflammatory effects through heme oxygenase-1 expression in murine macrophages. J. Pharmacol. Sci. 2011, 116, 283–295. [Google Scholar]
- Choi, A.M.; Alam, J. Heme oxygenase-1: Function, regulation, and implication of a novel stress-inducible protein in oxidant-induced lung injury. Am. J. Respir. Cell Mol. Biol. 1996, 15, 9–19. [Google Scholar] [CrossRef] [PubMed]
- MacMicking, J.D.; Nathan, C.; Hom, G.; Chartrain, N.; Fletcher, D.S.; Trumbauer, M.; Stevens, K.; Xie, Q.W.; Sokol, K.; Hutchinson, N.; et al. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell 1995, 81, 641–650. [Google Scholar]
- Pinho, B.R.; Sousa, C.; Valentão, P.; Andrade, P.B. Is nitric oxide decrease observed with naphthoquinones in LPS stimulated RAW264.7 macrophages a beneficial property? PLoS One 2011, 6, e24098. [Google Scholar]
- Coleman, J.W. Nitric oxide in immunity and inflammation. Int. Immunopharmacol. 2001, 1, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Ignarro, L.J. Nitric oxide as a unique signaling molecule in the vascular system: A historical overview. J. Physiol. Pharmacol. 2002, 53, 503–514. [Google Scholar] [PubMed]
- Sweet, M.J.; Hume, D.A. Endotoxin signal transduction in macrophages. J. Leukoc. Biol. 1996, 60, 8–26. [Google Scholar] [PubMed]
- Turini, M.E.; DuBois, R.N. Cyclooxygenase-2: A therapeutic target. Annu. Rev. Med. 2002, 53, 35–57. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Proinflammatory cytokines. Chest 2000, 118, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.Y.; Hung, T.M.; Bae, K.H.; Shin, E.M.; Zhou, H.Y.; Hong, Y.N.; Kang, S.S.; Kim, H.P.; Kim, Y.S. Anti-inflammatory effects of schisandrin isolated from the fruit of Schisandra chinensis Baill. Eur. J. Pharmacol. 2008, 591, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Magnani, M.; Crinelli, R.; Bianchi, M.; Antonelli, A. The ubiquitin-dependent proteolytic system and other potential targets for the modulation of nuclear factor-κB (NF-κB). Curr. Drug Targets 2000, 1, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.V.; Tan, A.S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): Subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. [Google Scholar] [PubMed]
- Titheradge, M.A. The enzymatic measurement of nitrate and nitrite. Meth. Mol. Biol. 1998, 100, 83–91. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-S.; Cui, X.; Lee, D.-S.; Ko, W.; Sohn, J.H.; Yim, J.H.; An, R.-B.; Kim, Y.-C.; Oh, H. Inhibitory Effects of Benzaldehyde Derivatives from the Marine Fungus Eurotium sp. SF-5989 on Inflammatory Mediators via the Induction of Heme Oxygenase-1 in Lipopolysaccharide-Stimulated RAW264.7 Macrophages. Int. J. Mol. Sci. 2014, 15, 23749-23765. https://doi.org/10.3390/ijms151223749
Kim K-S, Cui X, Lee D-S, Ko W, Sohn JH, Yim JH, An R-B, Kim Y-C, Oh H. Inhibitory Effects of Benzaldehyde Derivatives from the Marine Fungus Eurotium sp. SF-5989 on Inflammatory Mediators via the Induction of Heme Oxygenase-1 in Lipopolysaccharide-Stimulated RAW264.7 Macrophages. International Journal of Molecular Sciences. 2014; 15(12):23749-23765. https://doi.org/10.3390/ijms151223749
Chicago/Turabian StyleKim, Kyoung-Su, Xiang Cui, Dong-Sung Lee, Wonmin Ko, Jae Hak Sohn, Joung Han Yim, Ren-Bo An, Youn-Chul Kim, and Hyuncheol Oh. 2014. "Inhibitory Effects of Benzaldehyde Derivatives from the Marine Fungus Eurotium sp. SF-5989 on Inflammatory Mediators via the Induction of Heme Oxygenase-1 in Lipopolysaccharide-Stimulated RAW264.7 Macrophages" International Journal of Molecular Sciences 15, no. 12: 23749-23765. https://doi.org/10.3390/ijms151223749



