Molecular Genetic Variability of Commercial and Wild Accessions of Passion Fruit (Passiflora spp.) Targeting ex Situ Conservation and Breeding
Abstract
:1. Introduction
2. Results
2.1. Tests for Cross-Species Amplification
| Passiflora Species | P. cincinnata | P. edulis | P. setacea | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Loci (N) | % | Loci (N) | % | Loci (N) | % | Loci (N) | % | |
| P. alata Curtis | 19 | 76 | 29 | 91 | 44 | 85 | 92 | 84 |
| P. cincinnata Mast | – | – | 24 | 75 | 42 | 81 | 66 | 76 |
| P. edulis Sims | 12 | 48 | – | – | 46 | 88 | 58 | 75 |
| P. foetida L. | 14 | 56 | 22 | 69 | 35 | 67 | 71 | 65 |
| P. galbana Mast | 19 | 76 | 28 | 88 | 45 | 87 | 92 | 84 |
| P. gibertii N.E.Br | 16 | 64 | 21 | 66 | 36 | 69 | 73 | 67 |
| P. laurifolia L. | 12 | 48 | 12 | 38 | 26 | 50 | 50 | 46 |
| P. malacophylla Mast | 22 | 88 | 30 | 94 | 47 | 90 | 99 | 91 |
| P. morifolia Mast | 12 | 48 | 16 | 50 | 36 | 69 | 64 | 59 |
| P. rubra L. | 13 | 52 | 16 | 50 | 28 | 54 | 57 | 52 |
| P. setacea DC. | 6 | 24 | 21 | 66 | – | – | 27 | 47 |
| P. suberosa L. | 13 | 52 | 19 | 59 | 30 | 58 | 62 | 57 |
| P. tenuifila Killip | 15 | 60 | 22 | 69 | 38 | 73 | 75 | 69 |
| P. watsoniana Mast | 18 | 72 | 30 | 94 | 43 | 83 | 91 | 83 |
| Total | 15 | 59 | 22 | 70 | 38 | 73 | 70 | 68 |
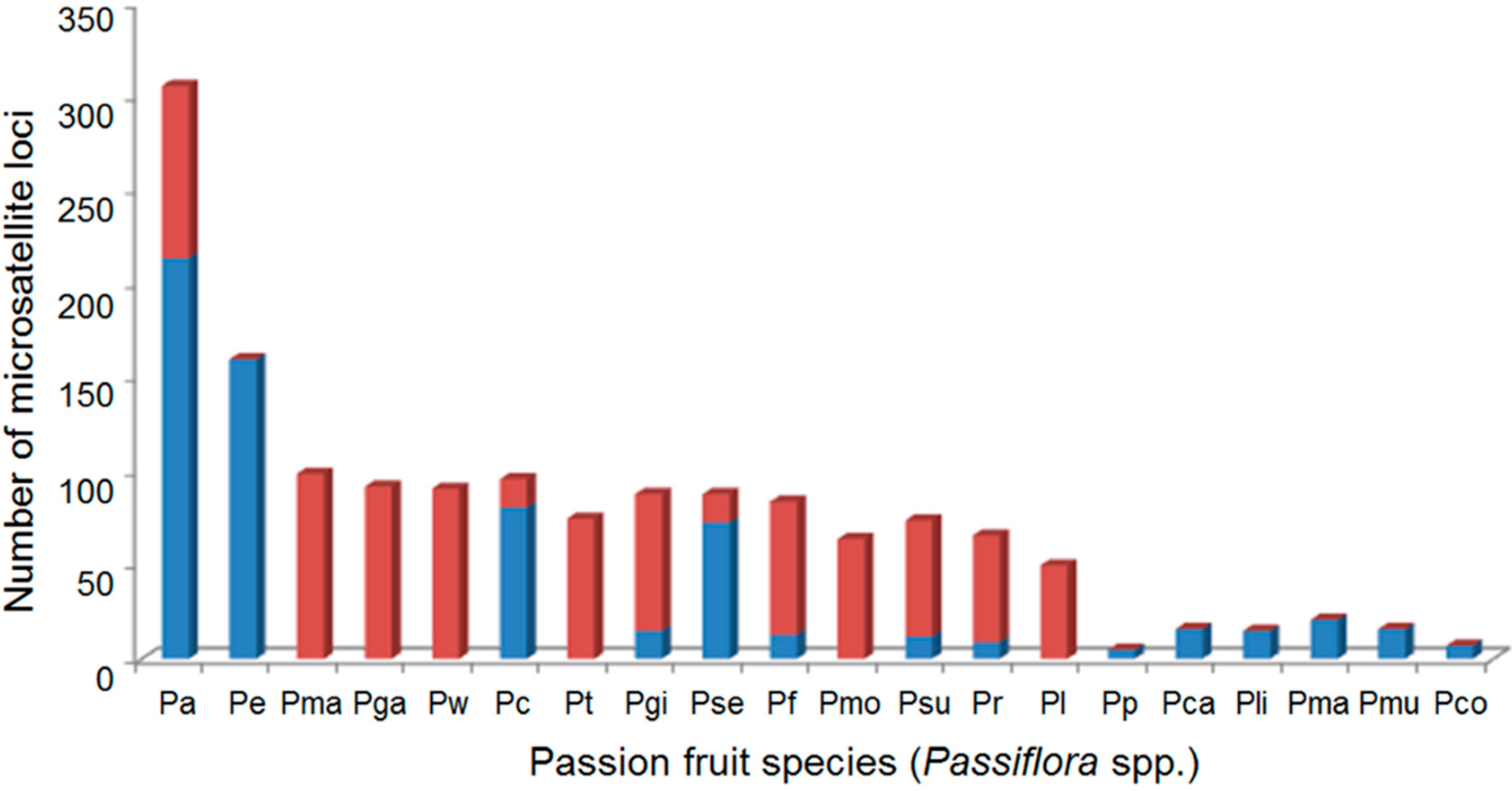
2.2. Polymorphism Analysis of Microsatellite Loci
| Passiflora Species | Values | Na | HO | HE | F | Fis |
|---|---|---|---|---|---|---|
| P. cincinnata | Minimum | 2 | 0.00 | 0.01 | −0.41 | −0.36 |
| Maximum | 16 | 0.92 | 0.89 | 1.00 | 0.66 | |
| Average | 6 | 0.42 | 0.52 | 0.17 | 0.15 | |
| Standard deviation | ±4.7 | ±0.2 | ±0.3 | ±0.3 | ±0.07 | |
| P. edulis | Minimum | 2 | 0.01 | 0.01 | −0.16 | −0.27 |
| Maximum | 18 | 0.77 | 0.86 | 0.38 | 0.31 | |
| Average | 6 | 0.43 | 0.50 | 0.13 | 0.05 | |
| Standard deviation | ±3.7 | ±0.2 | ±0.3 | ±0.1 | ±0.03 | |
| P. setacea | Minimum | 2 | 0.02 | 0.02 | −0.65 | −0.86 |
| Maximum | 6 | 0.82 | 0.69 | 0.87 | 0.68 | |
| Average | 3 | 0.25 | 0.36 | 0.28 | 0.08 | |
| Standard deviation | ±1.4 | ±0.2 | ±0.2 | ±0.4 | ±0.08 |
2.3. Genetic Structure and Diversity among Passion Fruit Accessions
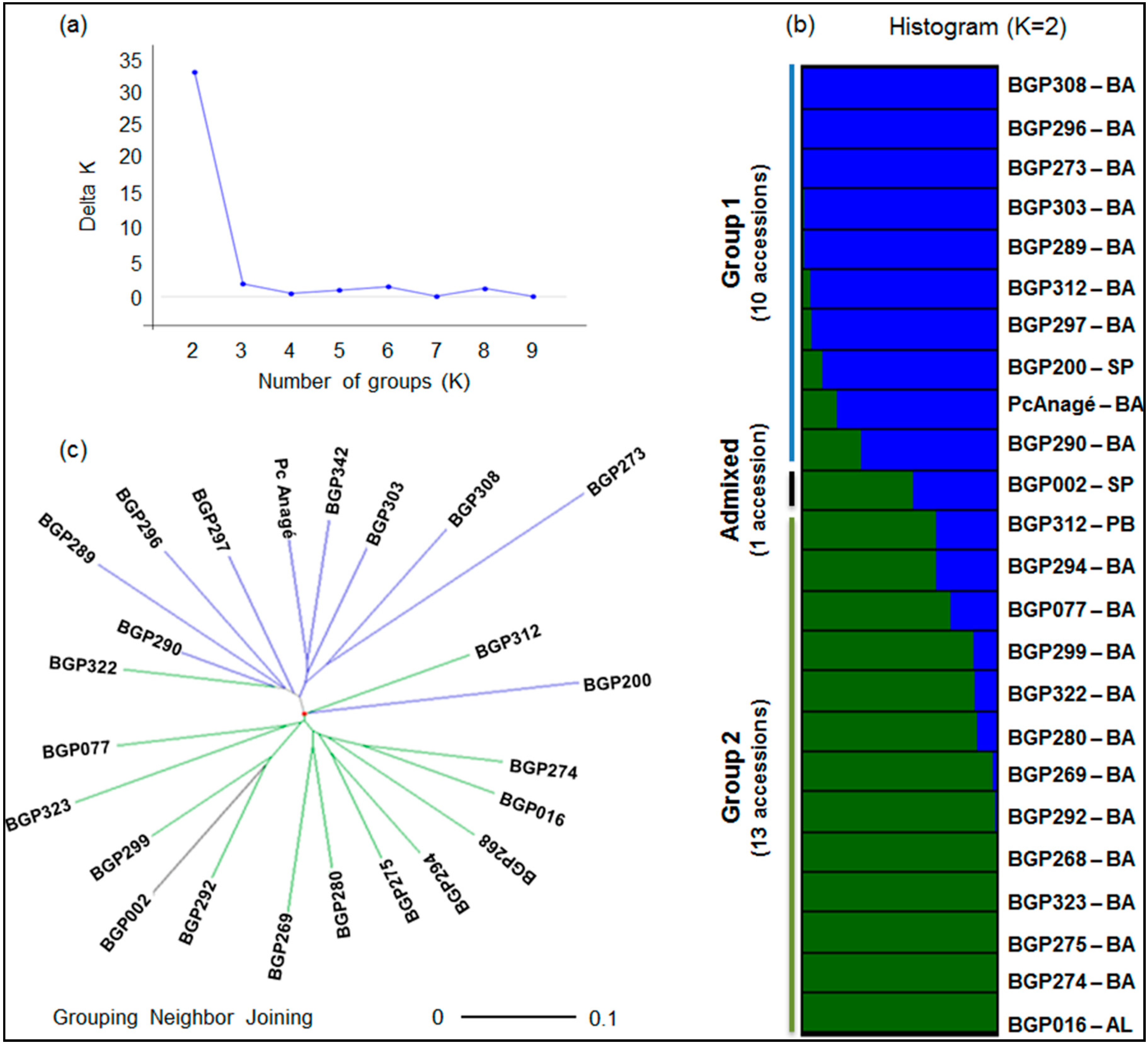

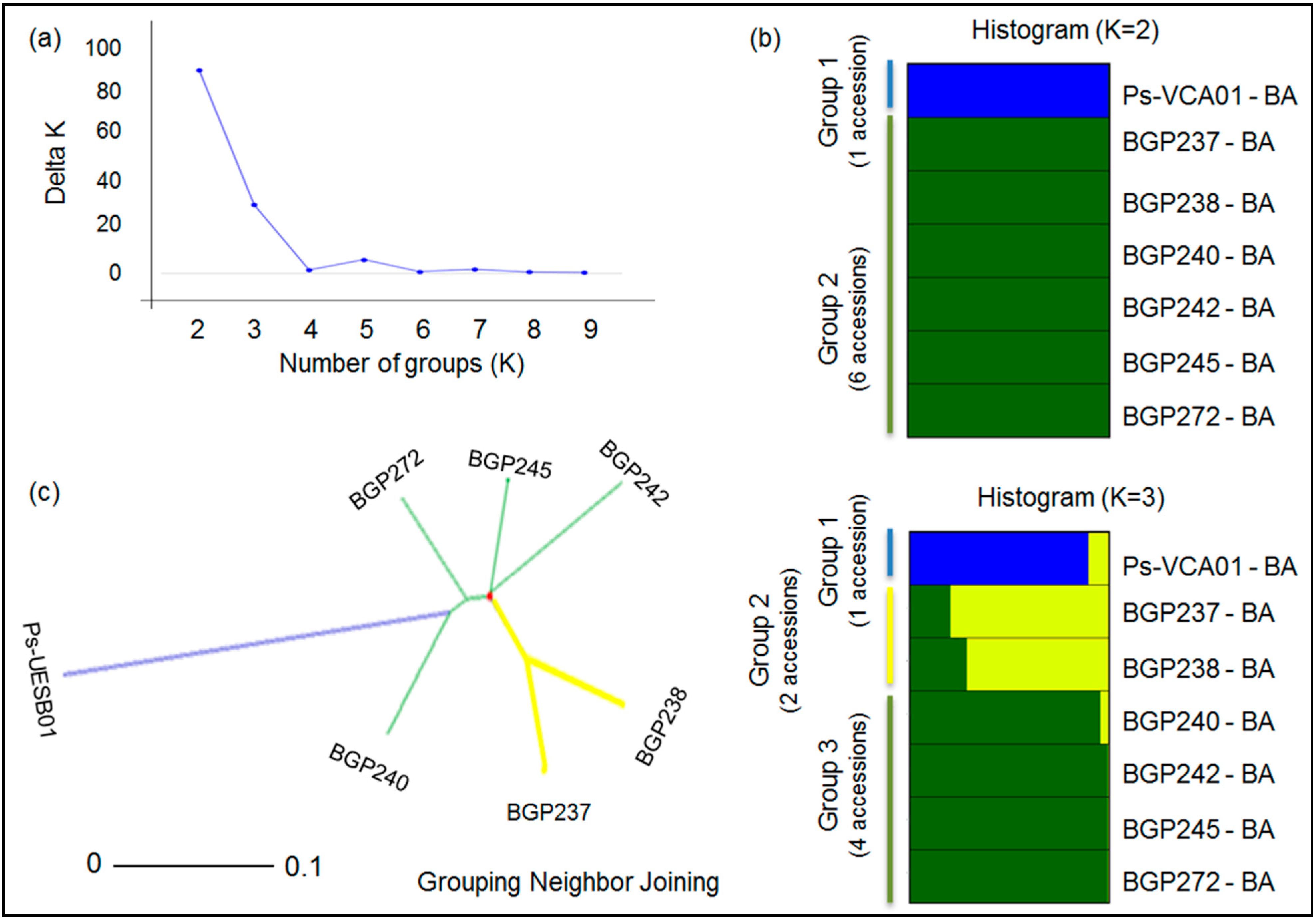
| Groups | P. cincinnata | P. edulis | P. setacea | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Nl | Na | NaR | Nl | Na | NaR | Nl | Na | NaR | |
| Group 1 | 10 | 27 | 70% | 2 | 2 | 100% | 17 | 24 | 25% |
| Group 2 | 13 | 29 | 69% | 1 | 1 | 100% | 2 | 2 | 100% |
| Group 3 | – | – | – | 8 | 13 | 31% | 3 | 4 | 50% |
| Group 4 | – | – | – | 6 | 8 | 75% | – | – | – |
| Group 5 | – | – | – | 6 | 7 | 75% | – | – | – |
| Total | 16 | 56 | 70% | 16 | 31 | 61% | 22 | 30 | 27% |
2.4. Estimates for the Formation of Core Collections

3. Discussion
3.1. Tests for Cross-Species Amplification
3.2. Polymorphism Analysis of Microsatellite Loci
3.3. Genetic Diversity and Structure among Passion Fruit Accessions
3.4. Formation of the Core Collection
4. Experimental Section
4.1. Germplasm Material and Genomic DNA Extraction
4.2. Analysis of Molecular Markers
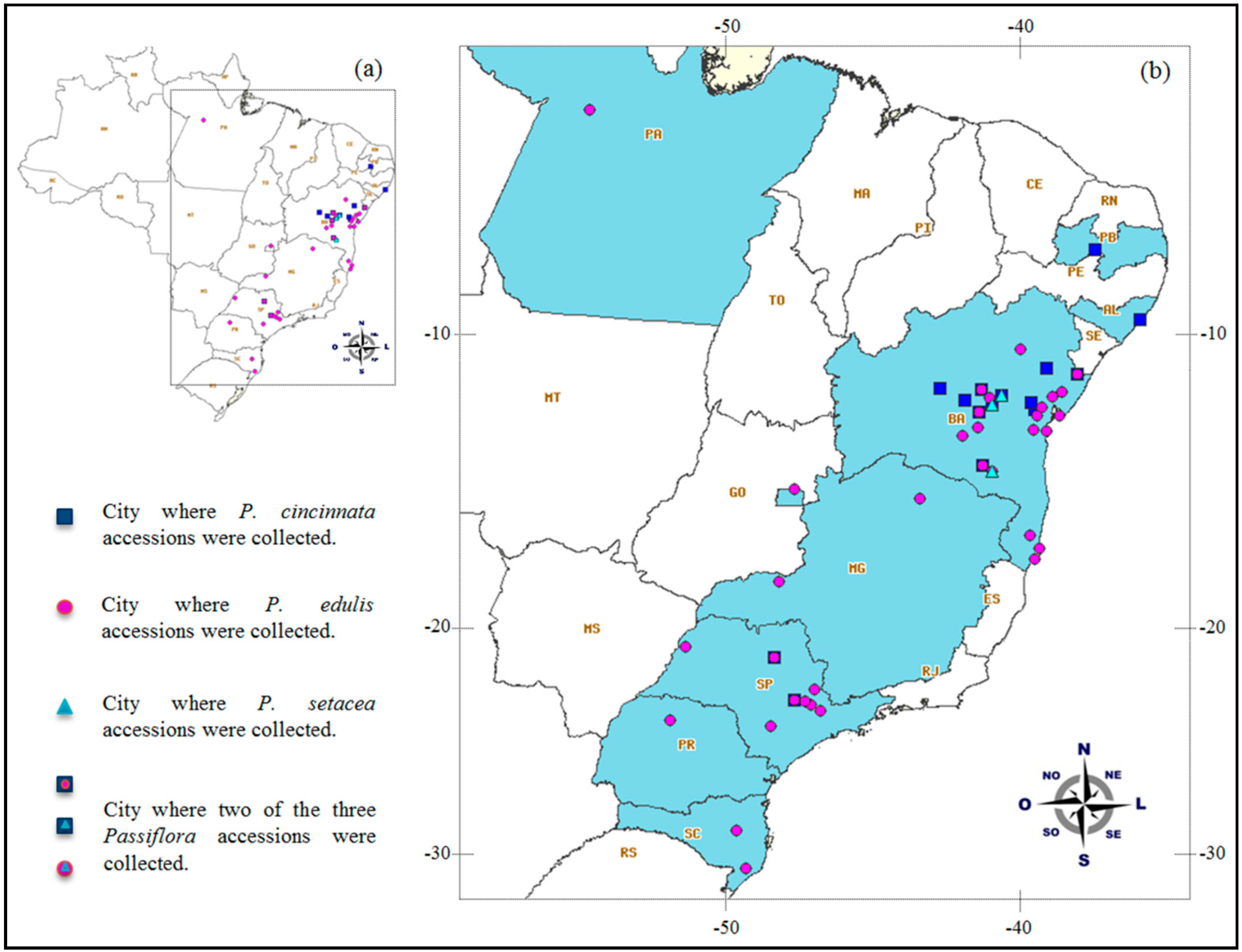
| Passiflora Species | Popular Name in Brazil | Natural Occurrence (Brazilian Macro-Regions) | Total Number of Accessions b | Total Number of Plants | Geographical Origin |
|---|---|---|---|---|---|
| P. cincinnata Mast | maracujá do mato/mochila | North—Northeast—Midwest—Southeast | 24 | 67 | Alagoas, Bahia, São Paulo, Paraíba |
| P. edulis Sims | maracujá azedo/amarelo | North—Northeast—Midwest—Southeast—South | 85 | 247 | Bahia, Distrito Federal, Minas Gerais, São Paulo, Santa Catarina, Pará, Paraná, Portugal, Venezuela |
| P. setacea DC. a | maracujá do sono/sururuca | Northeast—Midwest—Southeast | 7 | 50 | Bahia |
| P. alata Curtis a | maracujá doce | North—Northeast—Midwest—Southeast—South | 1 | 3 | Distrito Federal |
| P. gibertii N.E.Br | – | Midwest | 1 | 3 | Unavailable |
| P. laurifolia L. | maracujá laranja/perola | North—Northeast—Midwest | 1 | 3 | Unavailable |
| P. tenuifila Killip a | maracujá de cobra | South—Southeast | 1 | 3 | São Paulo |
| P. morifolia Mast | maracujá da capoeira/peludo | Midwest—Southeast—South | 1 | 3 | São Paulo |
| P. galbana Mast a | maracujá luxo | Northeast—Southeast | 1 | 3 | Unavailable |
| P. watsoniana Mast a | – | Northeast—Southeast | 1 | 3 | Unavailable |
| P. rubra L. | maracujá de estalo | Northeast—Southeast | 1 | 3 | São Paulo |
| P. suberosa L. | maracujá cortiça/maracujazinho | Northeast—Midwest—Southeast—South | 1 | 3 | São Paulo |
| P. foetida L. | – | North—Northeast—Midwest—Southeast—South | 1 | 3 | São Paulo |
| P. malacophylla Mast a | – | Northeast—Southeast | 1 | 3 | Unavailable |
4.3. Cross-Species Amplification and Characterization of Intraspecific Genetic Diversity
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Feuillet, C.; MacDougal, J.M. A new infrageneric classification of Passiflora L. (Passifloraceae). Passiflora 2004, 13, 34–38. [Google Scholar]
- Bernacci, L.C.; Meletti, L.M.M.; Soares-Scott, M.D.; Passos, I.R.S.; Junqueira, N.T.V. Espécies de maracujá: Caractreização e conservação da biodiversidade. In Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; Embrapa Cerrados: Planaltina, Brazil, 2005; pp. 558–586. (In Portuguese) [Google Scholar]
- Bernacci, L.C.; Cervi, A.C.; Milward-de-Azevedo, M.A.; Nunes, T.S.; Imig, D.C.; Mezzonato, A.C. Passifloraceae. Lista de espécies da flora do Brasil. In Jardim Botânico do Rio de Janeiro. Available online: http://floradobrasil.jbrj.gov.br/jabot/floradobrasil/FB182 (accessed on 20 January 2014). (In Portuguese)
- Ocampo, J.; d’Eeckenbrugge, J.C.; Jarvis, A. Distribution of the genus Passiflora. L. diversity in Colombia and its potential as an indicator for biodiversity management in the coffee growing zone. Diversity 2010, 2, 1158–1180. [Google Scholar] [CrossRef] [Green Version]
- Fajardo, D.; Angel, F.; Grum, M.; Tohme, J.; Lobo, M.; Roca, W.M.; Sanchez, I. Genetic variation analysis of the genus Passiflora. L. using RAPD markers. Euphytica 1998, 101, 341–347. [Google Scholar] [CrossRef]
- Viana, A.P.; Pereira, T.N.S.; Pereira, M.G.; Souza, M.M.; Maldonado, J.V.M.; Amaral Junior, A.T. Genetic diversity among yellow passion fruit commercial genotypes and among Passiflora. species using RAPD. Rev. Bras. Frutic. 2003, 25, 489–493. [Google Scholar] [CrossRef]
- Muschner, V.C.; Lorenz, A.P.; Cervi, A.C.; Bonatto, S.L.; Souza-Chies, T.T.; Salzano, F.M.; Freitas, L.B. A first molecular phylogenetic analysis of Passiflora. (Passifloracae). Am. J. Bot. 2003, 90, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Muschner, V.C.; Zamberlan, P.M.; Bonatto, S.L.; Freitas, L.B. Phylogeny, biogeography and divergence times in Passiflora. (Passifloraceae). Genet. Mol. Biol. 2012, 35, 1036–1043. [Google Scholar] [CrossRef]
- Pádua, J.G. Genetic Analysis of Species of the Genus Passiflora L. Based on Phylogenetic and Morphometric Approaches and on Microsatellite Markers. Ph.D. Thesis, Universidade Estadual de São Paulo, Piracicaba, São Paulo, Brazil, 16 September 2004. [Google Scholar]
- Killip, E.P. Publications field museum of natural history—Botanical series. In American Species of Passifloraceae; Field Museum of Natural History: London, UK, 1938; Volume 19, pp. 1–613. [Google Scholar]
- Escobar, L.K. A new subgenus and five new species in Passiflora. (Passifloraceae) from South America. Ann. Mo. Bot. Gard. 1989, 76, 877–885. [Google Scholar] [CrossRef]
- Hansen, A.K.; Gibert, L.E.; Simpson, B.B.; Downie, S.R.; Cervi, A.C.; Jansen, R.K. Phylogenetic relationships and chromosome number evolution in Passiflora. Syst. Bot. 2006, 31, 138–150. [Google Scholar] [CrossRef]
- Soares-Scott, M.D.; Meletti, L.M.M.; Bernacci, L.C.; Passos, I.R.S. Citogenética clássica e molecular em passifloras. In Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; Embrapa Cerrados: Planaltina, Brazil, 2005; pp. 210–240. (In Portuguese) [Google Scholar]
- Melo, N.F.; Guerra, M. Variability of the 5S and rDNA sites in Passiflora. L. with species with distinct base chromosome numbers. Ann. Bot. 2003, 92, 309–316. [Google Scholar] [CrossRef]
- Konta, E.M.; Almeida, M.R.; Amaral, C.L.; Darin, J.D.C.; Rosso, V.V.; Mercadante, A.Z.; Antunes, L.M.G.; Bianchi, M.L.P. Evaluation of the antihypertensive properties of yellow passion fruit pulp (Passiflora. edulis Sims f. flavicarpa Deg.) in spontaneously hypertensive rats. Phytother. Res. 2014, 28, 28–32. [Google Scholar]
- Costa, A.M.; Tupinambá, D.D.O. Maracujá e suas propriedade medicinais—Estado da arte. In Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; Embrapa Cerrados: Planaltina, Brazil, 2005; pp. 475–508. (In Portuguese) [Google Scholar]
- Dhawan, K.; Dhawan, S.; Sharma, A. Passiflora.: A review update. J. Ethnopharmacol. 2004, 94, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Faleiro, F.G.; Junqueira, N.T.V.; Braga, M.F.; Peixoto, J.R. Pré-melhoramento do maracujá. In Pré-Melhoramento de Plantas. Estado da Arte e Experiências de Sucesso; Lopes, M.A., Fávero, A.P., Ferreira, M.A.J.F., Faleiro, F.G., Folle, S.M., Guimarães, E.P., Eds.; Embrapa Informações Tecnologicas: Brasilia, Brazil, 2011; pp. 549–570. (In Portuguese) [Google Scholar]
- Zeraik, M.L.; Pereira, C.A.M.; Zuin, V.G.; Yariware, J.H. Passion fruit: A functional food? Braz. J. Pharmacogn. 2010, 20, 459–471. (In Portuguese) [Google Scholar] [CrossRef]
- Ferreira, F.R. Recursos genéticos de Passiflora. In Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; Embrapa Cerrados: Planaltina, Brazil, 2005; pp. 41–50. (In Portuguese) [Google Scholar]
- Souza, J.S.I.; Meletti, L.M.M. Maracujá: Espécies, Variedades, Cultivo; Fealq: Piracicaba, Brazil, 1997; p. 179. (In Portuguese) [Google Scholar]
- Santos, E.A. Improvment of Passifloras for Using Ornamentation Passiflora palmeri var. sublanceolata, Passiflora foetida var. foetida and Hybrids F1 Ornamental: Confirmation by RAPD, Genetic Parameters and Effects of Shading. Master Thesis, Universidade Estadual de Santa Cruz, Ilhéus, Bahia, Brazil, 10 July 2008. [Google Scholar]
- Peixoto, M. Problemas e perspectivas do maracujá ornamental. In Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; Embrapa Cerrados: Planaltina, Brazil, 2005; pp. 456–464. (In Portuguese) [Google Scholar]
- Bellon, G.; Faleiro, F.G.; Junqueira, K.P.; Junqueira, N.T.V.; Santos, E.C.; Braga, M.F.; Guimarães, C.T. Genetic variability of wild and commercial passion fruit (Passiflora edulis Sims) accessions using RAPD markers. Rev. Bras. Frutic. 2007, 29, 124–127. (In Portuguese) [Google Scholar] [CrossRef]
- IBGE—Diretoria de Pesquisas, Coordenação de Agropecuária, Produção Agrícola Municipal. 2012. Available online: http://www.ibge.gov.br/home/estatistica/economia/pam/2012/default_perm_xls.shtm (accessed on 14 March 2014). (In Portuguese)
- Meletti, L.M.M.; Soares-Scott, M.D.; Bernacci, L.C.; Passos, I.R.S. Melhoramento genético do maracujá: Passado e futuro. In Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; Embrapa Cerrados: Planaltina, Brazil, 2005; pp. 55–78. (In Portuguese) [Google Scholar]
- Melo, N.F.; Cervi, A.C.; Guerra, M. Kariology and citotaxonomy of the genus Passiflora. L. Plant Syst. Evol. Berl. 2001, 226, 68–84. [Google Scholar]
- Oliveira, E.J.; Soares, T.L.; Barbosa, C.J.; Santos Filho, H.P.; Jesus, O.N. Disease severity from passion fruit to identify sources of resistance in field conditions. Rev. Bras. Frutic. 2013, 35, 485–492. (In Portuguese) [Google Scholar] [CrossRef]
- Junqueira, N.T.V.; Anjos, J.R.N.; Silva, A.N.P.; Chaves, R.C.; Gomes, A.C. Reaction to diseases and yield of eleven cultivars of sour-passion fruit cultivated with no pesticides. Pesq. Agropec. Bras. 2003, 38, 1005–1010. [Google Scholar] [CrossRef]
- Junqueira, N.T.V.; Braga, M.F.; Faleiro, F.G.; Peixoto, J.R.; Bernacci, L.C. Potencial de espécies silvestres de maracujazeiro como fonte de resistência a de doenças. In Maracujá: Germoplasma e melhoramento genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; Embrapa Cerrados: Planaltina, Brazil, 2005; pp. 79–108. (In Portuguese) [Google Scholar]
- Junqueira, N.T.V.; Santos, E.C.; Junqueira, K.P.; Faleiro, F.G.; Bellon, G.; Braga, M.F. Physical and chemical characteristics and yield of Passiflora nitida Kunth accessions from north and central regions of Brazil. Rev. Bras. Frutic. 2010, 32, 791–797. (In Portuguese) [Google Scholar] [CrossRef]
- Cerqueira-Silva, C.B.M.; Cardoso-Silva, C.B.; Nonato, J.V.A.; Corrêa, R.X.; Oliveira, A.C. Genetic dissimilarity of “yellow” and “sleep” passion fruit accessions based on the fruits physical-chemical characteristics. Crop. Breed. Appl. Biotechnol. 2009, 9, 210–218. [Google Scholar] [CrossRef]
- Araújo, F.P.; Silva, N.; Queiroz, M.A. Genetic divergence among Passiflora. Cincinnata Mast accessions based on morphoagronomic descriptors. Rev. Bras. Frutic. 2008, 30, 723–730. [Google Scholar] [CrossRef]
- Meletti, L.M.M.; Bernacci, L.C.; Soares-Scott, M.D.; Azevedo Filho, J.A.; Martins, A.M. Genetic variability of morphological, agronomic and cytogenetics characters of sweet passion-fruit populations (Passiflora alata Curtis). Rev. Bras. Frutic. 2003, 25, 275–278. (In Portuguese) [Google Scholar] [CrossRef]
- Oliveira, J.C. Melhoramento Genético de P. edulis f. flavicarpa Deg. Visando Aumento de Produtividade. Ph.D. Thesis, Universidade Estadual de São Paulo, Jaboticabal, São Paulo, Brazil, 1980. [Google Scholar]
- Cerqueira-Silva, C.B.M.; Jesus, O.N.; Santos, E.S.L.; Corrêa, R.X.; Souza, A.P. Genetic breeding and diversity of the genus Passiflora.: Progress and perspectives in molecular and genetic studies. Int. J. Mol. Sci. 2014, 15, 14122–14152. [Google Scholar]
- Nass, L.L. Pré-melhoramento vegetal. In Pré-melhoramento de Plantas. Estado da Arte e Experiências de Sucesso; Lopes, M.A., Fávero, A.P., Ferreira, M.A.J.F., Faleiro, F.G., Folle, S.M., Guimarães, E.P., Eds.; Embrapa Informações Tecnologicas: Brasilia, Brazil, 2011; pp. 23–38. (In Portuguese) [Google Scholar]
- Lopes, M.A.; Fávero, A.P.; Faleiro, F.G. Expectativas e impactos do pré-melhoramento na gestão, no acesso e no uso de variabilidade genética vegetal. In Pré-melhoramento de plantas. Estado da arte e experiências de sucesso; Lopes, M.A., Fávero, A.P., Ferreira, M.A.J.F., Faleiro, F.G., Folle, S.M., Guimarães, E.P., Eds.; Embrapa Informações Tecnologicas: Brasília, 2011; pp. 85–98. (In Portuguese) [Google Scholar]
- Oliveira, E.J. Development of Microsatellite Markers and Their Use for the Generation and Integration of Genetic Maps of Yellow Passion Fruit (Passiflora edulis Sims f. flavicarpa Deg.) (Passiflora edulis Sims f. flavicarpa Deg.). Ph.D. Thesis, Universidade Estadual de São Paulo, Piracicaba, São Paulo, Brazil, 20 April 2006. [Google Scholar]
- Cerqueira-Silva, C.B.M.; Santos, E.S.L.; Souza, A.M.; Mori, G.M.; Oliveira, E.J.; Corrêa, R.X.; Souza, A.P. Development and characterization of microsatellite markers for the wild South American Passiflora. cincinnata (Passifloraceae). Am. J. Bot. 2012, 99, e170–e172. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira-Silva, C.B.M.; Santos, E.S.L.; Vieira, J.G.P.; Mori, G.M.; Jesus, O.N.; Correa, R.X.; Souza, A.P. New microsatellite markers for wild and commercial species of Passiflora. (Passifloraceae) and cross-amplification. Appl. Plant Sci. 2014, 2, 1–5. [Google Scholar]
- Wright, S. Evolution and the Genetics of Populations, Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Wetzel Wetzel, M.M.V.S.; Gimenes, M.A.; Pádua, J.G.; José, S.C.B.R.; Neto, L.G.P. Conservação de espécies silvestres com potencial de utilização em programas de pré-melhoramento na coleção base da Embrapa. In Pré-melhoramento de Plantas. Estado da Arte e Experiências de Sucesso; Lopes, M.A., Fávero, A.P., Ferreira, M.A.J.F., Faleiro, F.G., Folle, S.M., Guimarães, E.P., Eds.; Embrapa Informações Tecnologicas: Brasilia, Brazil, 2011; pp. 99–122. (In Portuguese) [Google Scholar]
- Ferreira, M.E.; Rangel, P.H.N. Aporte biotecnológico ao pré-melhoramento vegetal. In Pré-melhoramento de Plantas. Estado da arte e Experiências de Sucesso; Lopes, M.A., Fávero, A.P., Ferreira, M.A.J.F., Faleiro, F.G., Folle, S.M., Guimarães, E.P., Eds.; Embrapa Informações Tecnologicas: Brasília, 2011; pp. 59–84. (In Portuguese) [Google Scholar]
- Faleiro, F.G.; Junqueira, N.T.V.; Braga, M.F.; Bellon, G.; Peixoto, J.R. Diversidade genética de variedades comerciais de maracujazeiro-azedo com base em marcadores RAPD. In Reunião Técnica de Pesquisas em Maracujazeiro, 4; Planaltina: Brasília, Brazil, 2005; pp. 105–109. (In Portuguese) [Google Scholar]
- Pereira, M.G.; Pereira, T.N.S.; Costa, F. Marcadores moleculares no pré-melhoramento. In Marcadores Molecular; Borém, A., Caixeta, E.T., Eds.; Universidade Federal de Viçosa: Minas Gerais, Brazil, 2009; pp. 103–128. (In Portuguese) [Google Scholar]
- Reis, R.V.; Oliveira, E.J.; Viana, A.P.; Pereira, T.N.S.; Pereira, M.G.; Silva, M.G.M. Genetic diversity in recurrent selection of yellow passion fruit detected by microsatellites markers. Pesq. Agropec. Bras. 2011, 46, 51–57. (In Portuguese) [Google Scholar] [CrossRef]
- Reis, R.V.; Viana, A.P.; Oliveira, E.J.; Silva, M.G.M. Phenotypic and molecular selection of passion fruit progenies in the second cycle of recurrent selection. Crop. Breed. Appl. Biotechnol. 2012, 12, 17–24. [Google Scholar] [CrossRef]
- Oliveira, G.A.F.; Pádua, J.G.; Costa, J.L.; Jesus, O.N.; Carvalho, F.M.; Oliveira, E.J. Cross-species amplification of microsatellite loci developed for Passiflora edulis Sims. in related Passiflora. Species. Braz. Arch. Biol. Technol. 2013, 56, 785–792. [Google Scholar] [CrossRef]
- Paiva, C.L.; Viana, A.P.; Santos, E.A.; Freitas, J.C.O.; Silva, R.N.O.; Oliveira, E.J. Genetic variability assessment in the genus Passiflora by SSR markers. Chil. J. Agric. Res. 2014, 74, 355–360. [Google Scholar] [CrossRef]
- Barbará, T.; Palma-Silva, C.; Paggi, G.M.; Bered, F.; Fay, M.F.; Lexer, C. Cross-species transfer of nuclear microsatellite markers: Potential and limitations. Mol. Ecol. 2007, 16, 3759–3767. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira-Silva, C.B.M. Biometric, Genetic and Molecular Evaluations Targeting Conservation and Improvement of Passiflora spp. Master Dissertation, Universidade Estadual de Santa Cruz, Ilhéus, Bahia, Brazil, 13 February 2009. [Google Scholar]
- Penha, H.A.; Pereira, G.S.; Zucchi, M.I.; Diniz, A.L.; Vieira, M.L.C.; Flachowsky, H. Development of microsatellite markers in sweet passion fruit, and identification of length and conformation polymorphisms within repeat sequences. Plant Breed. 2013, 132, 731–735. [Google Scholar] [CrossRef]
- Cazé, A.L.R.; Kriedt, R.A.; Beheregaray, L.B.; Bonatto, S.L.; Freitas, L.B. Isolation and characterization of microsatellite markers for Passiflora contracta. Int. J. Mol. Sci. 2012, 13, 11343–11348. [Google Scholar] [CrossRef]
- Pádua, J.G.; Oliveira, E.J.; Zucchi, M.I.; Oliveira, G.C.X.; Camargo, L.E.A.; Vieira, M.L.C. Isolation and characterization of microsatellite markers from the sweet passion fruit (Passiflora alata Curtis: Passifloraceae). Mol. Ecol. Notes 2005, 5, 863–865. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Pádua, J.G.; Zucchi, M.I.; Camargo, L.E.A.; Fungaro, M.H.P.; Vieira, M.L.C. Development and characterization of microsatellite markers from the yellow passion fruit (Passiflora edulis f. flavicarpa). Mol. Ecol. Notes 2005, 5, 331–333. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Vieira, M.L.C.; Garcia, A.A.F.; Munhoz, C.E.F.; Margarido, G.R.A.; Consoli, L.; Matta, F.P.; Moraes, M.C. An integrated molecular map of yellow passion fruit based on simultaneous maximum-likelihood estimation of linkage and linkage phases. J. Am. Soc. Hortic. Sci. 2008, 133, 35–41. [Google Scholar]
- Santos, E.A.; Souza, M.M.; Abreu, P.P.; Conceição, L.D.H.C.S.; Araujo, I.S.; Viana, A.P.; Almeida, A.F.; Freitas, J.C.O. Confirmation and characterization of interspecific hybrids of Passiflora L. (Passifloraceae) for ornamental use. Euphytica 2012, 184, 389–399. [Google Scholar] [CrossRef]
- Pereira, G.S.; Nunes, E.S.; Laperuta, L.D.C.; Braga, M.F.; Penha, H.A.; Diniz, A.L.; Munhoz, C.F.; Gazaffi, R.; Garcia, A.A.F.; Vieira, M.L.C. Molecular polymorphism and linkage analysis in sweet passion fruit, an outcrossing species. Ann. Appl. Biol. 2013, 162, 347–361. [Google Scholar] [CrossRef]
- Pereira, G.S. Development of SSR, M-AFLP and SNP Markers for Linkage Map Integration of Passiflora alata Curtis. Master Dissertation, Universidade Estadual de São Paulo, Piracicaba, São Paulo, Brazil, 31 January 2011. [Google Scholar]
- Ortiz, D.C.; Bohórquez, A.; Duque, M.C.; Tohme, J.; Cuéllar, D.; Vásquez, T.M. Evaluating purple passion fruit (Passiflora edulis Sims f. edulis) genetic variability in individuals from commercial plantations in Colombia. Genet. Resour. Crop. Evol. 2012, 59, 1089–1099. [Google Scholar]
- Bruckner, C.H.; Suassuna, T.M.F.; Rego, M.M.; Nunes, E.S. Auto-incompatibilidade do maracujá: implicações no melhoramento genético. In Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; Embrapa Cerrados: Planaltina, Brazil, 2005; pp. 475–506. (In Portuguese) [Google Scholar]
- Santos, L.F.; Oliveira, E.J.; Silva, A.S.; Carvalho, F.M.; Costa, J.L.; Pádua, J.G. ISSR markers as a tool for the assessment of genetic diversity in Passiflora. Biochem. Genet. 2011, 49, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Ganga, R.M.D.; Ruggiero, C.; Lemos, E.G.M.; Grili, G.V.G.; Gonçalves, M.M.; Chagas, E.A.; Wickert, E. Genetic diversity in yellow passion fruit utilizing fAFLP molecular markers. Rev. Bras. Frutic. 2004, 26, 494–498. (In Portuguese) [Google Scholar] [CrossRef]
- Cerqueira-Silva, C.B.M.; Santos, E.S.L.; Conceição, L.D.H.C.S.; Cardoso-Silva, C.B.; Pereira, A.S.; Oliveira, A.C.; Corrêa, R.X. Short communication genetic variation in a wild population of the sleep passion fruit (Passiflora setacea) based on molecular markers. Genet. Mol. Res. 2012, 11, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira-Silva, C.B.M.; Conceição, L.D.H.C.S.; Santos, E.S.L.; Cardoso-Silva, C.B.; Pereira, A.S.; Oliveira, A.C.; Corrêia, R.X. Genetic variability in wild genotypes of Passiflora cincinnata based on RAPD markers. Genet. Mol. Res. 2010, 9, 2421–2428. [Google Scholar] [CrossRef] [PubMed]
- Lista de Espécies da Flora do Brasil. In Jardim Botânico do Rio de Janeiro. Available online: http://floradobrasil.jbrj.gov.br (accessed on 19 August 2014). (In Portuguese)
- Cerqueira-Silva, C.B.M.; Conceição, L.D.H.C.S.; Cardoso-Silva, C.B.; Pereira, A.S.; Santos, E.S.L.; Oliveira, A.C.; Correa, R.X. Genetic diversity of yellow passion fruit (Passiflora edulis Sims) based on RAPD markers. Crop. Breed. Appl. Biotechnol. 2010, 10, 154–159. [Google Scholar] [CrossRef]
- Freitas, J.P.X.; Oliveira, E.J.; Neto, A.J.C.; Santos, L.R. Evaluation of genetic resources of yellow passion fruit. Pesq. Agropec. Bras. 2011, 46, 1013–1020. [Google Scholar] [CrossRef]
- Cerqueira-Silva, C.B.M.; Jesus, O.N.; Oliveira, E.J.; Santos, E.S.L.; Souza, A.P. Characterization and selection of passion fruit (yellow and purple) accessions based on molecular markers and disease reactions for use in breeding programs. Euphytica 2014, 199. [Google Scholar] [CrossRef]
- Bernacci, L.C.; Soares-Scott, M.D.; Junqueira, N.T.V.; Passos, I.R.S.; Meletti, L.M.M. Passiflora edulis Sims: The correct taxonomic way to cite the yellow passion fruit (and of others colors). Rev. Bras. Frutic. 2008, 30, 566–576. [Google Scholar] [CrossRef]
- Santos-Garcia, M.O.; Toledo-Silva, G.; Sassaki, R.P.; Ferreira, T.H.; Resende, R.M.S.; Chiari, L.; Karia, C.T.; Carvalho, M.A.; Faleiro, F.G.; Zucchi, M.I.; et al. Using genetic diversity information to establish core collections of Stylosanthes capitata and Stylosanthes macrocephala. Genet. Mol. Biol. 2012, 35, 847–861. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.H.D.; Grace, J.P.; Speer, S.S. Designation of a “core” collection of perennial Glycine. Soybean Genet. Newsl. 1987, 14, 59–70. [Google Scholar]
- Van Hintum, T.J.L. The general methodology for creating a core collection. In Core Collections for Today and Tomorrow; Johnson, R.C., Hodgkin, T., Eds.; International Plant Genetic Resources Institute: Rome, Italy, 1999; pp. 10–17. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Creste, S.; Tulmann Neto, A.; Figueira, A. Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol. Biol. Rep. 2001, 19, 299–306. [Google Scholar] [CrossRef]
- SpeciesMapper Tool. Available online: http://splink.cria.org.br/ (accessed on 19 August 2014).
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S. Genetic Data Analysis: Methods for Discrete Population Genetic Data; Sinauer Associates, Inc.: Sunderland, MA, USA, 1990. [Google Scholar]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Miller, M.P. Tools for Population Genetics Analyses (TFPGA) 1.3: A Windows Program for the Analysis of Allozyme and Molecular Population Genetic Data. 1997. Available online: http://herb.bio.nau.edul~miller/tfpga.htm (accessed on 14 March 2014).
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/ (accessed on 14 March 2014).
- Cipriani, G.; Spadotto, A.; Jurman, I.; Gaspero, G.; Crespan, M.; Meneghetti, S.; Frare, E.; Vignani, R.; Cresti, M.; Morgante, M.; et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 2010, 121, 1569–1585. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerqueira-Silva, C.B.M.; Santos, E.S.L.; Jesus, O.N.; Vieira, J.G.P.; Mori, G.M.; Corrêa, R.X.; Souza, A.P. Molecular Genetic Variability of Commercial and Wild Accessions of Passion Fruit (Passiflora spp.) Targeting ex Situ Conservation and Breeding. Int. J. Mol. Sci. 2014, 15, 22933-22959. https://doi.org/10.3390/ijms151222933
Cerqueira-Silva CBM, Santos ESL, Jesus ON, Vieira JGP, Mori GM, Corrêa RX, Souza AP. Molecular Genetic Variability of Commercial and Wild Accessions of Passion Fruit (Passiflora spp.) Targeting ex Situ Conservation and Breeding. International Journal of Molecular Sciences. 2014; 15(12):22933-22959. https://doi.org/10.3390/ijms151222933
Chicago/Turabian StyleCerqueira-Silva, Carlos Bernard M., Elisa S. L. Santos, Onildo N. Jesus, João G. P. Vieira, Gustavo M. Mori, Ronan X. Corrêa, and Anete P. Souza. 2014. "Molecular Genetic Variability of Commercial and Wild Accessions of Passion Fruit (Passiflora spp.) Targeting ex Situ Conservation and Breeding" International Journal of Molecular Sciences 15, no. 12: 22933-22959. https://doi.org/10.3390/ijms151222933






