Inducible Expression of Mu-Class Glutathione S-Transferases Is Associated with Fenpropathrin Resistance in Tetranychus cinnabarinus
Abstract
:1. Introduction
2. Results and Discussion
2.1. Bioassay Results
| Strains | LC50 (mg/L) 95% CI | LC30 (mg/L) 95% CI | Slope (±SE) | RR |
|---|---|---|---|---|
| SS | 606.98 (490.80–727.67) | 280.63 (188.04–364.23) | 1.57 ± 0.21 | – |
| FR | 61,581.93 (49,641.61–75,068.16) | 27,710.96 (18,492.14–36,037.43) | 1.51 ± 0.21 | 101 |
2.2. Sequencing and Annotation of GSTs from T. cinnabarinus
| Gene | D2 | D3 | D4 | D5 |
|---|---|---|---|---|
| D1 | 76.85% | 75.93% | 79.63% | 46.91% |
| D2 | – | 85.65% | 74.07% | 45.66% |
| D3 | – | – | 71.76% | 43.83% |
| D4 | – | – | – | 44.50% |
| Gene | M2 | M3 | M4 | M5 | M6 | M7 | M8 |
|---|---|---|---|---|---|---|---|
| M1 | 37.5% | 38.63% | 39.65% | 37.89% | 40.17% | 39.47% | 41.41% |
| M2 | – | 66.38% | 46.61% | 40.44% | 45.78% | 40.09% | 39.65% |
| M3 | – | – | 40.68% | 39.41% | 41.18% | 41.53% | 38.98% |
| M4 | – | – | – | 59.03% | 47.60% | 73.57% | 69.60% |
| M5 | – | – | – | – | 40.03% | 63.00% | 60.79% |
| M6 | – | – | – | – | – | 44.10% | 41.92% |
| M7 | – | – | – | – | – | – | 72.96% |
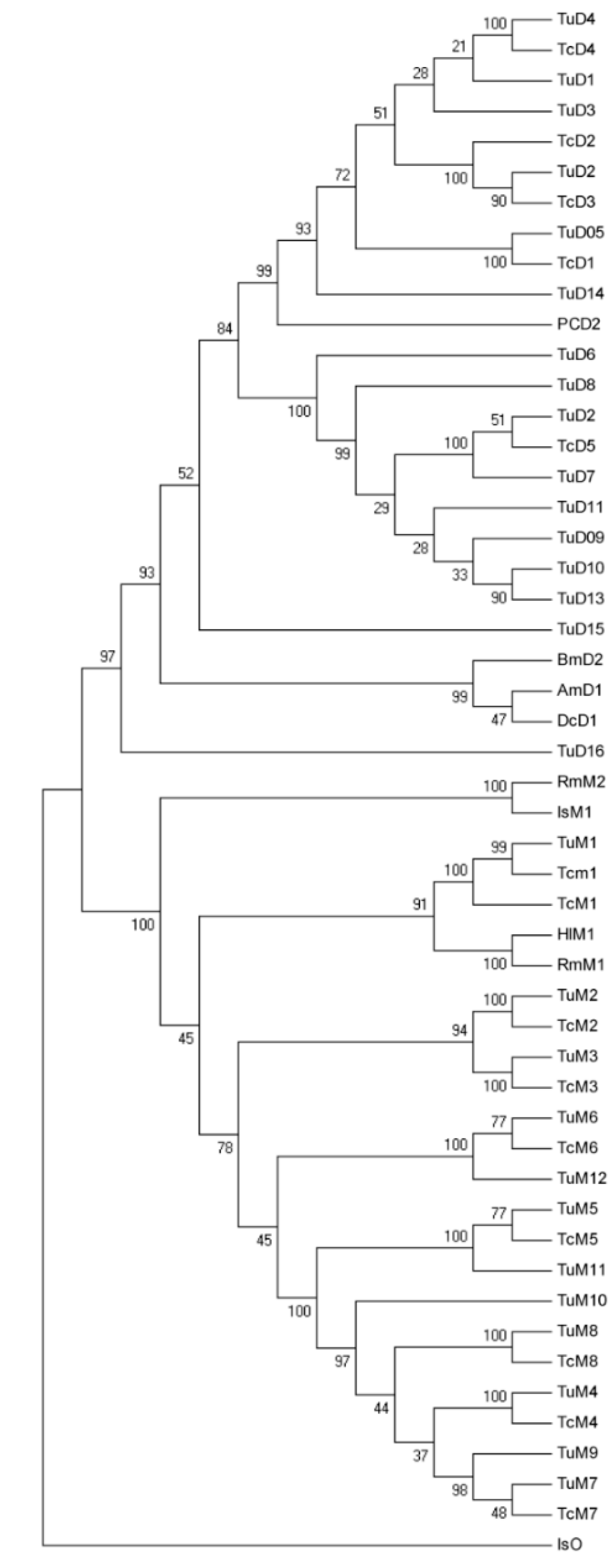
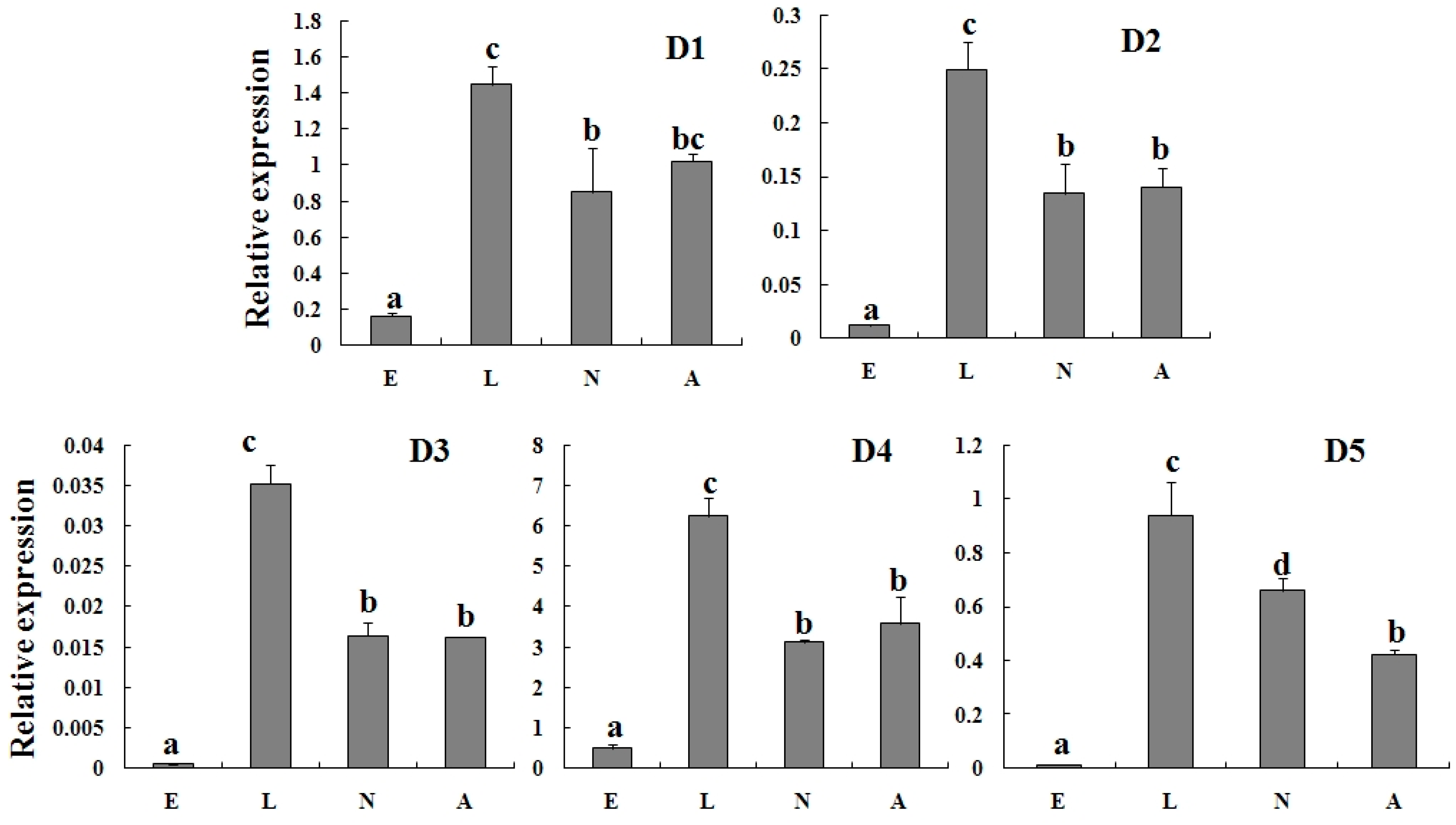
2.3. Specific Expression of TcGSTs in Different Developmental Stages
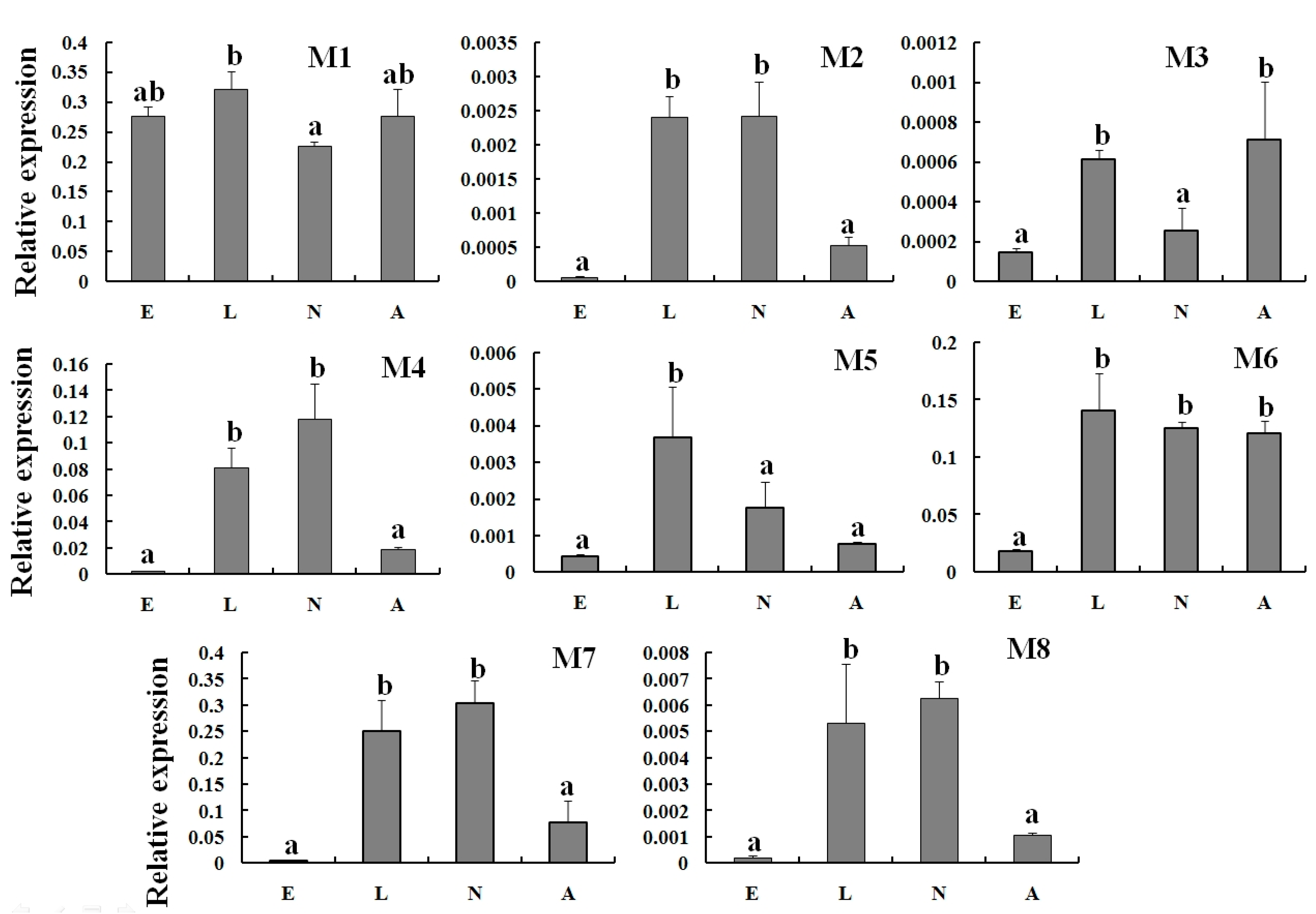
2.4. Expression Profiles of TcGSTs between Fenpropathrin-Resistant (FR) and Susceptible Strains (SS)


2.5. Expression Reaction of TCGSTs to Fenpropathrin Stimulation



3. Experimental Section
3.1. Mites
3.2. Bioassay and Acaricide Stimulation
3.3. RNA Extraction and cDNA Synthesis
3.4. Cloning of Full-Length GST cDNA and Mutation Alignment
3.5. Phylogenetic Analysis
3.6. Real-Time Quantitative PCR and Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guo, F.; Zhang, Z.; Zhao, Z. Pesticide resistance of Tetranychus cinnabarinus (Acari: tetranychidae) in China: A review. Syst. Appl. Acarol. 1998, 3, 3–7. [Google Scholar]
- Feng, Y.N.; Yan, J.; Sun, W.; Zhao, S.; Lu, W.C.; Li, M.; He, L. Transcription and induction profiles of two esterase genes in susceptible and acaricide-resistant Tetranychus cinnabarinus. Pestic. Biochem. Physiol. 2011, 100, 70–73. [Google Scholar]
- De Mendonca, R.S.; Navia, D.; Diniz, I.R.; Auger, P.; Navajas, M. A critical review on some closely related species of Tetranychus sensustricto (Acari: tetranychidae) in the public DNA sequences databases. Exp. Appl. Acarol. 2011, 55, 1–23. [Google Scholar]
- Li, T.; Chen, X.L.; Hong, X.Y. Population genetic structure of Tetranychus urticae and its sibling species Tetranychus cinnabaribus (Acari: tetranychidae) in China as inferred from microsatellite data. Ann. Entomol. Soc. Am. 2009, 102, 674–683. [Google Scholar]
- Auger, P.; Migeon, A.; Ueckermann, E.A.; Tiedt, L.; Navajas, M. Evidence for synonymy between Tetranychus urticae and Tetranychus cinnabaribus (Acari, prostigmata, Tetranychidae): Review and new data. Acarologia 2013, 53, 383–415. [Google Scholar]
- Kwon, D.H.; Yoon, K.S.; Clark, J.M.; Lee, S.H. A point mutation in a glutamate-gated chloride channel confers abamectin resistance in the two-spotted spider mite, Tetranychus urticae Koch. Insect Mol. Biol. 2010, 19, 583–591. [Google Scholar]
- Feng, Y.N.; Zhao, S.; Sun, W.; Li, M.; Lu, W.; He, L. The sodium channel gene in Tetranychus cinnabarinus (Boisduval): Identification and expression analysis of a mutation associated with pyrethroid resistance. Pest Manag. Sci. 2011, 67, 904–912. [Google Scholar]
- Khajehali, J.; van Leeuwen, T.; Grispou, M.; Morou, E.; Alout, H.; Weill, M.; Tirry, L.; Vontas, J.; Tsagkarakou, A. Acetylcholinesterase point mutations in European strains of Tetranychus urticae (Acari: tetranychidae) resistant to organophosphates. Pest Manag. Sci. 2010, 66, 220–228. [Google Scholar]
- Van Leeuwen, T.; Vontas, J.; Tsagkarakou, A.; Dermauw, W.; Tirry, L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem. Mol. Biol. 2010, 40, 563–572. [Google Scholar]
- Takahashi, N.; Nakagawa, H.; Sasama, Y.; Ikemi, N. Development of a new acaricide, cyflumetofen. J. Pestic. Sci. 2012, 37, 263–264. [Google Scholar]
- He, L.; Zhao, Z.M.; Deng, X.P.; Wang, J.J.; Liu, H. Resistance risk assessment: Realized heritability of resistance to methrin, abamectin, pyridaben and their mixtures in the spider mite, Tetranychus cinnabarinus. Int. J. Pest Manag. 2003, 49, 271–274. [Google Scholar]
- Chen, Q.S.; Zhao, S.; Zou, J.; Shi, L.; He, L. Monitoring of acaricide resistance in Tetranychus cinnabarinus. Chin. J. Appl. Entomol. 2012, 49, 364–369. [Google Scholar]
- Tsagkarakou, A.; van Leeuwen, T.; Khajehali, J.; Ilias, A.; Grispou, M.; Williamson, M.S.; Tirry, L.; Vontas, J. Identification of pyrethroid resistance associated mutations in the para sodium channel of the two-spotted spider mite Tetranychus urticae (Acari: tetranychidae). Insect Mol. Biol. 2009, 18, 583–593. [Google Scholar]
- Xu, Z.F.; Shi, L.; Feng, Y.N.; He, L. The molecular marker of kdr against fenpropathrin in Tetranychus cinnabarinus. J. Econ. Entomol. 2014, 106, 2457–2466. [Google Scholar]
- He, L.; Xue, C.H.; Wang, J.J.; Li, M.; Lu, W.C.; Zhao, Z.M. Resistance selection and biochemical mechanism of resistance to two Acaricides in Tetranychus cinnabarinus (Boiduval). Pestic. Biochem. Physiol. 2009, 93, 47–52. [Google Scholar]
- Enayati, A.A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005, 14, 3–8. [Google Scholar]
- Qin, G.; Liu, T.; Guo, Y.; Zhang, X.; Ma, E.; Zhang, J. Effects of chlorpyrifos on glutathione S-transferase in migratory locust, Locusta migratoria. Pestic. Biochem. Physiol. 2014, 109, 1–5. [Google Scholar]
- Hu, F.; Dou, W.; Wang, J.J.; Jia, F.X.; Wang, J.J. Multiple glutathione S-transferase genes: Identification and expression in oriental fruit fly, Bactrocera dorsalis. Pest Manag. Sci. 2013, 70, 295–303. [Google Scholar]
- Grbić, M.; van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouzé, P.; Grbić, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.T.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar]
- Armstrong, R. Glutathione S-transferases: Reaction mechanism, structure, and function. Chem. Res. Toxicol. 1991, 4, 131–140. [Google Scholar]
- Wu, B.; Dong, D. Human cytosolic glutathione transferases: Structure, function, and drug discovery. Trends Pharmacol. Sci. 2012, 33, 656–668. [Google Scholar]
- Liao, C.Y.; Zhang, K.; Niu, J.Z.; Ding, T.B.; Zhong, R.; Xia, W.K.; Dou, W.; Wang, J.J. Identification and characterization of seven glutathione S-transferase genes from citrus red mite, Panonychus citri (McGregor). Int. J. Mol. Sci. 2013, 14, 24255–24270. [Google Scholar]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar]
- Xu, Z.F.; Zhu, W.Y.; Liu, Y. Analysis of insecticide resistance-related genes of the carmine spider mite Tetranychus cinnabarinus based on a de novo assembled transcriptome. PLoS One 2014, 9, e94779. [Google Scholar]
- Zou, F.M.; Lou, D.S.; Zhu, Y.H.; Wang, S.P.; Jin, B.R.; Gui, Z.Z. Expression profiles of glutathione S-transferase genes in larval midgut of Bombyxmori exposed to insect hormones. Mol. Biol. Rep. 2011, 38, 639–647. [Google Scholar]
- Qin, G.; Jia, M.; Liu, T.; Xuan, T.; Zhu, Y.K.; Guo, Y.; Ma, E.; Zhang, J. Identification and characterisation of ten glutathione S-transferase genes from oriental migratory locust, Locusta migratoria manilensis (Meyen). Pest Manag. Sci. 2011, 67, 697–704. [Google Scholar]
- Li, X.C.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar]
- Wu, S.W.; Yang, Y.H.; Yuan, G.R.; Campbell, P.M.; Teese, M.G.; Russell, R.J.; Oakeshott, J.G.; Wu, Y. Overexpressed esterases in a fenvalerate resistant strain of the cotton bollworm, Helicoverpa armigera. Insect Biochem. Mol. Biol. 2011, 41, 14–21. [Google Scholar]
- Yang, T.; Liu, N. Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS One 2011, 6, e29418. [Google Scholar]
- Lumjuan, N.K.; Rajatileka, S.; Changsom, D.; Wicheera, J.; Leelapata, P.; Prapanthadaraa, L.; Somboonb, P.; Lycettc, G.; Ranson, H. The role of the Aedes aegypti epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect Biochem. Mol. Biol. 2012, 41, 203–209. [Google Scholar]
- Kliot, A.; Ghanim, M. Fitness costs associated with insecticide resistance. Pest Manag. Sci. 2012, 68, 1431–1437. [Google Scholar]
- Chung, H.; Boey, A.; Lumb, C.; Willoughby, L. Induction of a detoxification gene in Drosophila melanogaster requires an interaction between tissue specific enhancers and a novel cis-regulatory element. Insect Biochem. Mol. Biol. 2011, 41, 863–871. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar]
- Sun, W.; Jin, Y.; He, L.; Lu, W.C.; Li, M. Suitable reference gene selection for different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. J. Insect Sci. 2010, 10, 208. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative Ct method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, G.-M.; Shi, L.; Xu, Z.-F.; He, L. Inducible Expression of Mu-Class Glutathione S-Transferases Is Associated with Fenpropathrin Resistance in Tetranychus cinnabarinus. Int. J. Mol. Sci. 2014, 15, 22626-22641. https://doi.org/10.3390/ijms151222626
Shen G-M, Shi L, Xu Z-F, He L. Inducible Expression of Mu-Class Glutathione S-Transferases Is Associated with Fenpropathrin Resistance in Tetranychus cinnabarinus. International Journal of Molecular Sciences. 2014; 15(12):22626-22641. https://doi.org/10.3390/ijms151222626
Chicago/Turabian StyleShen, Guang-Mao, Li Shi, Zhi-Feng Xu, and Lin He. 2014. "Inducible Expression of Mu-Class Glutathione S-Transferases Is Associated with Fenpropathrin Resistance in Tetranychus cinnabarinus" International Journal of Molecular Sciences 15, no. 12: 22626-22641. https://doi.org/10.3390/ijms151222626




